[med-svn] [Git][med-team/python-bioframe][upstream] New upstream version 0.4.1
Nilesh Patra (@nilesh)
gitlab at salsa.debian.org
Sun May 7 09:40:18 BST 2023
Nilesh Patra pushed to branch upstream at Debian Med / python-bioframe
Commits:
0476c7c3 by Nilesh Patra at 2023-05-07T14:07:56+05:30
New upstream version 0.4.1
- - - - -
25 changed files:
- CHANGES.md
- README.md
- bioframe/_version.py
- bioframe/core/arrops.py
- bioframe/core/specs.py
- bioframe/extras.py
- bioframe/io/fileops.py
- bioframe/ops.py
- bioframe/sandbox/clients.py
- + docs/figs/bioframe_closest.pdf
- + docs/figs/closest0.png
- + docs/figs/closest1.png
- + docs/figs/closest2.png
- + docs/figs/closest3.png
- docs/guide-definitions.rst
- docs/guide-intervalops.md
- docs/guide-recipes.md
- + docs/guide-specifications.rst
- docs/index.rst
- docs/tutorials/tutorial_assign_motifs_to_peaks.ipynb
- + docs/tutorials/tutorial_assign_peaks_to_genes.ipynb
- tests/test_core_specs.py
- tests/test_extras.py
- tests/test_ops.py
- + tests/test_ops_select.py
Changes:
=====================================
CHANGES.md
=====================================
@@ -1,5 +1,28 @@
# Release notes
+## [v0.4.1](https://github.com/open2c/bioframe/compare/v0.4.0...v0.4.1)
+
+Date 2023-04-22
+
+Bug fixes:
+* Fix bug introduced in the last release in `select` and `select_*` query interval semantics. Results of select are now consistent with the query interval being interpreted as half-open, closed on the left.
+
+
+## [v0.4.0](https://github.com/open2c/bioframe/compare/v0.3.3...v0.4.0)
+
+Date 2023-03-23
+
+API changes:
+* New strand-aware directionality options for `closest()` via `direction_col` #129.
+* New index-based range query selectors on single bioframes to complement `select()` #128:
+ * `select_mask()` returns boolean indices corresponding to intervals that overlap the query region
+ * `select_indices()` returns integer indices corresponding to intervals that overlap the query region
+ * `select_labels()` returns pandas label indices corresponding to intervals that overlap the query region
+
+Bug fixes:
+* Import fixes in sandbox
+* Relax bioframe validator to permit using same column as start and end (e.g. point variants).
+
## [v0.3.3](https://github.com/open2c/bioframe/compare/v0.3.2...v0.3.3)
Date: 2022-02-28
=====================================
README.md
=====================================
@@ -11,9 +11,12 @@ The philosophy underlying bioframe is to enable flexible operations: instead of
Bioframe implements a variety of genomic interval operations directly on dataframes. Bioframe also includes functions for loading diverse genomic data formats, and performing operations on special classes of genomic intervals, including chromosome arms and fixed size bins.
-Read the [docs](https://bioframe.readthedocs.io/en/latest/), including the [guide](https://bioframe.readthedocs.io/en/latest/guide-intervalops.html).
+Read the [docs](https://bioframe.readthedocs.io/en/latest/), including the [guide](https://bioframe.readthedocs.io/en/latest/guide-intervalops.html), as well as the [bioframe preprint](https://doi.org/10.1101/2022.02.16.480748) for more information.
+
+If you use ***bioframe*** in your work, please cite:
+*Bioframe: Operations on Genomic Intervals in Pandas Dataframes*. Open2C, Nezar Abdennur, Geoffrey Fudenberg, Ilya Flyamer, Aleksandra A. Galitsyna, Anton Goloborodko, Maxim Imakaev, Sergey V. Venev.
+bioRxiv 2022.02.16.480748; doi: https://doi.org/10.1101/2022.02.16.480748
-If you use ***bioframe*** in your work, please cite via its zenodo DOI 10.5281/zenodo.5703622
## Installation
The following are required before installing bioframe:
=====================================
bioframe/_version.py
=====================================
@@ -1 +1 @@
-__version__ = "0.3.3"
+__version__ = "0.4.1"
=====================================
bioframe/core/arrops.py
=====================================
@@ -484,12 +484,12 @@ def complement_intervals(
def _closest_intervals_nooverlap(
- starts1, ends1, starts2, ends2, tie_arr=None, k_upstream=1, k_downstream=1
+ starts1, ends1, starts2, ends2, direction, tie_arr=None, k=1
):
"""
For every interval in set 1, return the indices of k closest intervals
- from set 2. Overlapping intervals from set 2 are not reported, unless they
- overlap by a single point.
+ from set 2 to the left from the interval (with smaller coordinate).
+ Overlapping intervals from set 2 are not reported, unless they overlap by a single point.
Parameters
----------
@@ -497,20 +497,23 @@ def _closest_intervals_nooverlap(
Interval coordinates. Warning: if provided as pandas.Series, indices
will be ignored.
+ direction : str ("left" or "right")
+ Orientation of closest interval search
+
tie_arr : numpy.ndarray or None
Extra data describing intervals in set 2 to break ties when multiple intervals
are located at the same distance. An interval with the *lowest* value is
selected.
- k_upstream, k_downstream : int
- The number of upstream and downstream neighbors to report.
+ k : int
+ The number of neighbors to report.
Returns
-------
- upstream_ids, downstream_ids: numpy.ndarray
- Two Nx2 arrays containing the indices of pairs of closest intervals,
- reported separately for the downstream and upstream neighbors. The two columns
- are the inteval ids from set 1, ids of the closest intevals from set 2.
+ ids: numpy.ndarray
+ One Nx2 array containing the indices of pairs of closest intervals,
+ reported for the neighbors in specified direction (by genomic coordinate).
+ The two columns are the inteval ids from set 1, ids of the closest intevals from set 2.
"""
@@ -530,12 +533,9 @@ def _closest_intervals_nooverlap(
n1 = starts1.shape[0]
n2 = starts2.shape[0]
- upstream_ids, downstream_ids = (
- np.zeros((0, 2), dtype=int),
- np.zeros((0, 2), dtype=int),
- )
+ ids = np.zeros((0, 2), dtype=int)
- if k_upstream > 0:
+ if k > 0 and direction=="left":
if tie_arr is None:
ends2_sort_order = np.argsort(ends2)
else:
@@ -544,26 +544,26 @@ def _closest_intervals_nooverlap(
ids2_endsorted = np.arange(0, n2)[ends2_sort_order]
ends2_sorted = ends2[ends2_sort_order]
- upstream_closest_endidx = np.searchsorted(ends2_sorted, starts1, "right")
- upstream_closest_startidx = np.maximum(upstream_closest_endidx - k_upstream, 0)
+ left_closest_endidx = np.searchsorted(ends2_sorted, starts1, "right")
+ left_closest_startidx = np.maximum(left_closest_endidx - k, 0)
int1_ids = np.repeat(
- np.arange(n1), upstream_closest_endidx - upstream_closest_startidx
+ np.arange(n1), left_closest_endidx - left_closest_startidx
)
int2_sorted_ids = arange_multi(
- upstream_closest_startidx, upstream_closest_endidx
+ left_closest_startidx, left_closest_endidx
)
- upstream_ids = np.vstack(
+ ids = np.vstack(
[
int1_ids,
ids2_endsorted[int2_sorted_ids],
# ends2_sorted[int2_sorted_ids] - starts1[int1_ids],
- # arange_multi(upstream_closest_startidx - upstream_closest_endidx, 0)
+ # arange_multi(left_closest_startidx - left_closest_endidx, 0)
]
).T
- if k_downstream > 0:
+ elif k > 0 and direction=="right":
if tie_arr is None:
starts2_sort_order = np.argsort(starts2)
else:
@@ -572,28 +572,29 @@ def _closest_intervals_nooverlap(
ids2_startsorted = np.arange(0, n2)[starts2_sort_order]
starts2_sorted = starts2[starts2_sort_order]
- downstream_closest_startidx = np.searchsorted(starts2_sorted, ends1, "left")
- downstream_closest_endidx = np.minimum(
- downstream_closest_startidx + k_downstream, n2
+ right_closest_startidx = np.searchsorted(starts2_sorted, ends1, "left")
+ right_closest_endidx = np.minimum(
+ right_closest_startidx + k, n2
)
int1_ids = np.repeat(
- np.arange(n1), downstream_closest_endidx - downstream_closest_startidx
+ np.arange(n1), right_closest_endidx - right_closest_startidx
)
int2_sorted_ids = arange_multi(
- downstream_closest_startidx, downstream_closest_endidx
+ right_closest_startidx, right_closest_endidx
)
- downstream_ids = np.vstack(
+ ids = np.vstack(
[
int1_ids,
ids2_startsorted[int2_sorted_ids],
# starts2_sorted[int2_sorted_ids] - ends1[int1_ids],
- # arange_multi(1, downstream_closest_endidx -
- # downstream_closest_startidx + 1)
+ # arange_multi(1, right_closest_endidx -
+ # right_closest_startidx + 1)
]
).T
- return upstream_ids, downstream_ids
+
+ return ids
def closest_intervals(
@@ -606,6 +607,7 @@ def closest_intervals(
ignore_overlaps=False,
ignore_upstream=False,
ignore_downstream=False,
+ direction=None
):
"""
For every interval in set 1, return the indices of k closest intervals from set 2.
@@ -631,6 +633,9 @@ def closest_intervals(
ignore_upstream, ignore_downstream : bool
If True, ignore set 2 intervals upstream/downstream of set 1 intervals.
+ direction : numpy.ndarray with dtype bool or None
+ Strand vector to define the upstream/downstream orientation of the intervals.
+
Returns
-------
closest_ids : numpy.ndarray
@@ -640,6 +645,7 @@ def closest_intervals(
"""
+ # Get overlapping intervals:
if ignore_overlaps:
overlap_ids = np.zeros((0, 2), dtype=int)
elif (starts2 is None) and (ends2 is None):
@@ -649,24 +655,66 @@ def closest_intervals(
else:
overlap_ids = overlap_intervals(starts1, ends1, starts2, ends2)
- upstream_ids, downstream_ids = _closest_intervals_nooverlap(
- starts1,
- ends1,
+ # Get non-overlapping intervals:
+ n = len(starts1)
+ all_ids = np.arange(n)
+
+ # + directed intervals
+ ids_left_upstream = _closest_intervals_nooverlap(
+ starts1[direction],
+ ends1[direction],
+ starts2,
+ ends2,
+ direction="left",
+ tie_arr=tie_arr,
+ k=0 if ignore_upstream else k
+ )
+ ids_right_downstream = _closest_intervals_nooverlap(
+ starts1[direction],
+ ends1[direction],
starts2,
ends2,
- tie_arr,
- k_upstream=0 if ignore_upstream else k,
- k_downstream=0 if ignore_downstream else k,
+ direction="right",
+ tie_arr=tie_arr,
+ k=0 if ignore_downstream else k
)
+ # - directed intervals
+ ids_right_upstream = _closest_intervals_nooverlap(
+ starts1[~direction],
+ ends1[~direction],
+ starts2,
+ ends2,
+ direction="right",
+ tie_arr=tie_arr,
+ k=0 if ignore_upstream else k
+ )
+ ids_left_downstream = _closest_intervals_nooverlap(
+ starts1[~direction],
+ ends1[~direction],
+ starts2,
+ ends2,
+ direction="left",
+ tie_arr=tie_arr,
+ k=0 if ignore_downstream else k
+ )
+
+ # Reconstruct original indexes (b/c we split regions by direction above)
+ ids_left_upstream[:, 0] = all_ids[direction][ids_left_upstream[:, 0]]
+ ids_right_downstream[:, 0] = all_ids[direction][ids_right_downstream[:, 0]]
+ ids_left_downstream[:, 0] = all_ids[~direction][ids_left_downstream[:, 0]]
+ ids_right_upstream[:, 0] = all_ids[~direction][ids_right_upstream[:, 0]]
+
+ left_ids = np.concatenate([ids_left_upstream, ids_left_downstream])
+ right_ids = np.concatenate([ids_right_upstream, ids_right_downstream])
# Increase the distance by 1 to distinguish between overlapping
# and non-overlapping set 2 intervals.
- upstream_dists = starts1[upstream_ids[:, 0]] - ends2[upstream_ids[:, 1]] + 1
- downstream_dists = starts2[downstream_ids[:, 1]] - ends1[downstream_ids[:, 0]] + 1
+ left_dists = starts1[left_ids[:, 0]] - ends2[left_ids[:, 1]] + 1
+ right_dists = starts2[right_ids[:, 1]] - ends1[right_ids[:, 0]] + 1
- closest_ids = np.vstack([upstream_ids, downstream_ids, overlap_ids])
+ closest_ids = np.vstack([left_ids, right_ids, overlap_ids])
closest_dists = np.concatenate(
- [upstream_dists, downstream_dists, np.zeros(overlap_ids.shape[0])]
+ [left_dists, right_dists, np.zeros(overlap_ids.shape[0])]
)
# Sort by distance to set 1 intervals and, if present, by the tie-breaking
=====================================
bioframe/core/specs.py
=====================================
@@ -58,7 +58,7 @@ class update_default_colnames:
_rc["colnames"] = self._old_colnames
-def _verify_columns(df, colnames, return_as_bool=False):
+def _verify_columns(df, colnames, unique_cols=False, return_as_bool=False):
"""
Raises ValueError if columns with colnames are not present in dataframe df.
@@ -78,8 +78,9 @@ def _verify_columns(df, colnames, return_as_bool=False):
return False
raise ValueError("df is not a dataframe")
- if len(set(colnames)) < len(colnames):
- raise ValueError("column names must be unique")
+ if unique_cols:
+ if len(set(colnames)) < len(colnames):
+ raise ValueError("column names must be unique")
if not set(colnames).issubset(df.columns):
if return_as_bool:
=====================================
bioframe/extras.py
=====================================
@@ -11,6 +11,7 @@ __all__ = [
"digest",
"frac_mapped",
"frac_gc",
+ "seq_gc",
"frac_gene_coverage",
"pair_by_distance",
]
@@ -76,7 +77,7 @@ def make_chromarms(
sk1, ek1 = "start", "end"
elif len(cols_chroms) == 3:
ck1, sk1, ek1 = cols_chroms
- _verify_columns(df_chroms, [ck1, sk1, ek1])
+ _verify_columns(df_chroms, [ck1, sk1, ek1], unique_cols=True)
if any((df_chroms[sk1].values != 0)):
raise ValueError("all values in starts column must be zero")
else:
@@ -278,7 +279,7 @@ def frac_gc(df, fasta_records, mapped_only=True, return_input=True):
Returns
-------
df_mapped : pd.DataFrame
- Original dataframe with new column 'frac_mapped' appended.
+ Original dataframe with new column 'GC' appended.
"""
if not set(df["chrom"].values).issubset(set(fasta_records.keys())):
@@ -297,16 +298,7 @@ def frac_gc(df, fasta_records, mapped_only=True, return_input=True):
gc = []
for _, bin in chrom_group.iterrows():
s = seq[bin.start : bin.end]
- g = s.count("G")
- g += s.count("g")
- c = s.count("C")
- c += s.count("c")
- nbases = len(s)
- if mapped_only:
- n = s.count("N")
- n += s.count("n")
- nbases -= n
- gc.append((g + c) / nbases if nbases > 0 else np.nan)
+ gc.append(seq_gc(s, mapped_only=mapped_only))
return gc
out = df.groupby("chrom", sort=False).apply(_each)
@@ -320,9 +312,42 @@ def frac_gc(df, fasta_records, mapped_only=True, return_input=True):
return pd.Series(data=np.concatenate(out), index=df.index).rename("GC")
+def seq_gc(seq, mapped_only=True):
+ """
+ Calculate the fraction of GC basepairs for a string of nucleotides.
+
+ Parameters
+ ----------
+ seq : str
+ Basepair input
+
+ mapped_only: bool
+ if True, ignore 'N' in the sequence for calculation.
+ if True and there are no mapped base-pairs, return np.nan.
+
+ Returns
+ -------
+ gc : float
+ calculated gc content.
+
+ """
+ if not type(seq) == str:
+ raise ValueError("reformat input sequence as a str")
+ g = seq.count("G")
+ g += seq.count("g")
+ c = seq.count("C")
+ c += seq.count("c")
+ nbases = len(seq)
+ if mapped_only:
+ n = seq.count("N")
+ n += seq.count("n")
+ nbases -= n
+ return (g + c) / nbases if nbases > 0 else np.nan
+
+
def frac_gene_coverage(df, ucsc_mrna):
"""
- Calculate number and fraction of overlaps by predicted and verified
+ Calculate number and fraction of overlaps by predicted and verified
RNA isoforms for a set of intervals stored in a dataframe.
Parameters
=====================================
bioframe/io/fileops.py
=====================================
@@ -5,6 +5,9 @@ import tempfile
import json
import io
+import os
+import shutil
+
import numpy as np
import pandas as pd
@@ -488,7 +491,7 @@ def read_bigbed(path, chrom, start=None, end=None, engine="auto"):
return df
-def to_bigwig(df, chromsizes, outpath, value_field=None):
+def to_bigwig(df, chromsizes, outpath, value_field=None, path_to_binary=None):
"""
Save a bedGraph-like dataframe as a binary BigWig track.
@@ -504,8 +507,34 @@ def to_bigwig(df, chromsizes, outpath, value_field=None):
value_field : str, optional
Select the column label of the data frame to generate the track. Default
is to use the fourth column.
+ path_to_binary : str, optional
+ Provide system path to the bedGraphToBigWig binary.
"""
+
+ if path_to_binary is None:
+ cmd = "bedGraphToBigWig"
+ try:
+ assert shutil.which(cmd) is not None
+ except Exception as e:
+ raise ValueError(
+ "bedGraphToBigWig is not present in the current environment. "
+ "Pass it as 'path_to_binary' parameter to bioframe.to_bigwig or "
+ "install it with, for example, conda install -y -c bioconda ucsc-bedgraphtobigwig "
+ )
+ elif path_to_binary.endswith("bedGraphToBigWig"):
+ if not os.path.isfile(path_to_binary) and os.access(path_to_binary, os.X_OK):
+ raise ValueError(
+ f"bedGraphToBigWig is absent in the provided path or cannot be executed: {path_to_binary}. "
+ )
+ cmd = path_to_binary
+ else:
+ cmd = os.path.join(path_to_binary, "bedGraphToBigWig")
+ if not os.path.isfile(cmd) and os.access(cmd, os.X_OK):
+ raise ValueError(
+ f"bedGraphToBigWig is absent in the provided path or cannot be executed: {path_to_binary}. "
+ )
+
is_bedgraph = True
for col in ["chrom", "start", "end"]:
if col not in df.columns:
@@ -527,7 +556,7 @@ def to_bigwig(df, chromsizes, outpath, value_field=None):
bg = bg.sort_values(["chrom", "start", "end"])
with tempfile.NamedTemporaryFile(suffix=".bg") as f, tempfile.NamedTemporaryFile(
- "wt", suffix=".chrom.sizes"
+ "wt", suffix=".chrom.sizes"
) as cs:
chromsizes.to_csv(cs, sep="\t", header=False)
@@ -538,14 +567,14 @@ def to_bigwig(df, chromsizes, outpath, value_field=None):
)
p = subprocess.run(
- ["bedGraphToBigWig", f.name, cs.name, outpath],
+ [cmd, f.name, cs.name, outpath],
stdout=subprocess.PIPE,
stderr=subprocess.PIPE,
)
return p
-def to_bigbed(df, chromsizes, outpath, schema="bed6"):
+def to_bigbed(df, chromsizes, outpath, schema="bed6", path_to_binary=None):
"""
Save a bedGraph-like dataframe as a binary BigWig track.
@@ -561,8 +590,34 @@ def to_bigbed(df, chromsizes, outpath, schema="bed6"):
value_field : str, optional
Select the column label of the data frame to generate the track. Default
is to use the fourth column.
+ path_to_binary : str, optional
+ Provide system path to the bedGraphToBigWig binary.
"""
+
+ if path_to_binary is None:
+ cmd = "bedToBigBed"
+ try:
+ assert shutil.which(cmd) is not None
+ except Exception as e:
+ raise ValueError(
+ "bedToBigBed is not present in the current environment. "
+ "Pass it as 'path_to_binary' parameter to bioframe.to_bigbed or "
+ "install it with, for example, conda install -y -c bioconda ucsc-bedtobigbed "
+ )
+ elif path_to_binary.endswith("bedToBigBed"):
+ if not os.path.isfile(path_to_binary) and os.access(path_to_binary, os.X_OK):
+ raise ValueError(
+ f"bedToBigBed is absent in the provided path or cannot be executed: {path_to_binary}. "
+ )
+ cmd = path_to_binary
+ else:
+ cmd = os.path.join(path_to_binary, "bedGraphToBigWig")
+ if not os.path.isfile(cmd) and os.access(cmd, os.X_OK):
+ raise ValueError(
+ f"bedToBigBed is absent in the provided path or cannot be executed: {path_to_binary}. "
+ )
+
is_bed6 = True
for col in ["chrom", "start", "end", "name", "score", "strand"]:
if col not in df.columns:
@@ -590,7 +645,7 @@ def to_bigbed(df, chromsizes, outpath, schema="bed6"):
)
p = subprocess.run(
- ["bedToBigBed", "-type={}".format(schema), f.name, cs.name, outpath],
+ [cmd, "-type={}".format(schema), f.name, cs.name, outpath],
stdout=subprocess.PIPE,
stderr=subprocess.PIPE,
)
=====================================
bioframe/ops.py
=====================================
@@ -11,6 +11,9 @@ from .core import checks
__all__ = [
"select",
+ "select_mask",
+ "select_indices",
+ "select_labels",
"expand",
"overlap",
"cluster",
@@ -27,19 +30,17 @@ __all__ = [
]
-def select(df, region, cols=None):
+def select_mask(df, region, cols=None):
"""
- Return all genomic intervals in a dataframe that overlap a genomic region.
+ Return boolean mask for all genomic intervals that overlap a query range.
Parameters
----------
df : pandas.DataFrame
region : str or tuple
- The genomic region to select from the dataframe.
- UCSC-style genomic region string, or Triple (chrom, start, end),
- where ``start`` or ``end`` may be ``None``. See :func:`.core.stringops.parse_region()`
- for more information on region formatting.
+ The genomic region to select from the dataframe in UCSC-style genomic
+ region string, or triple (chrom, start, end).
cols : (str, str, str) or None
The names of columns containing the chromosome, start and end of the
@@ -47,21 +48,107 @@ def select(df, region, cols=None):
Returns
-------
- df : pandas.DataFrame
-
+ Boolean array of shape (len(df),)
"""
-
ck, sk, ek = _get_default_colnames() if cols is None else cols
- checks.is_bedframe(df, raise_errors=True, cols=[ck, sk, ek])
+ _verify_columns(df, [ck, sk, ek])
chrom, start, end = parse_region(region)
+
if chrom is None:
raise ValueError("no chromosome detected, check region input")
- if (start is not None) and (end is not None):
- inds = (df[ck] == chrom) & (df[sk] < end) & (df[ek] > start)
+
+ if start is None:
+ mask = df[ck] == chrom
else:
- inds = df[ck] == chrom
- return df[inds]
+ if end is None:
+ end = np.inf
+ mask = (df[ck] == chrom) & (
+ ((df[sk] < end) & (df[ek] > start)) |
+ ((df[sk] == df[ek]) & (df[sk] == start)) # include points at query start
+ )
+ return mask.to_numpy()
+
+
+def select_indices(df, region, cols=None):
+ """
+ Return integer indices of all genomic intervals that overlap a query range.
+
+ Parameters
+ ----------
+ df : pandas.DataFrame
+
+ region : str or tuple
+ The genomic region to select from the dataframe in UCSC-style genomic
+ region string, or triple (chrom, start, end).
+
+ cols : (str, str, str) or None
+ The names of columns containing the chromosome, start and end of the
+ genomic intervals. The default values are 'chrom', 'start', 'end'.
+
+ Returns
+ -------
+ 1D array of int
+ """
+ return np.nonzero(select_mask(df, region, cols))[0]
+
+
+def select_labels(df, region, cols=None):
+ """
+ Return pandas Index labels of all genomic intervals that overlap a query
+ range.
+
+ Parameters
+ ----------
+ df : pandas.DataFrame
+
+ region : str or tuple
+ The genomic region to select from the dataframe in UCSC-style genomic
+ region string, or triple (chrom, start, end).
+
+ cols : (str, str, str) or None
+ The names of columns containing the chromosome, start and end of the
+ genomic intervals. The default values are 'chrom', 'start', 'end'.
+
+ Returns
+ -------
+ pandas.Index
+ """
+ return df.index[select_mask(df, region, cols)]
+
+
+def select(df, region, cols=None):
+ """
+ Return all genomic intervals in a dataframe that overlap a genomic region.
+
+ Parameters
+ ----------
+ df : pandas.DataFrame
+
+ region : str or tuple
+ The genomic region to select from the dataframe in UCSC-style genomic
+ region string, or triple (chrom, start, end).
+
+ cols : (str, str, str) or None
+ The names of columns containing the chromosome, start and end of the
+ genomic intervals. The default values are 'chrom', 'start', 'end'.
+
+ Returns
+ -------
+ df : pandas.DataFrame
+
+ Notes
+ -----
+ See :func:`.core.stringops.parse_region()` for more information on region
+ formatting.
+
+ See also
+ --------
+ :func:`select_mask`
+ :func:`select_indices`
+ :func:`select_labels`
+ """
+ return df.loc[select_mask(df, region, cols)]
def expand(df, pad=None, scale=None, side="both", cols=None):
@@ -816,6 +903,7 @@ def _closest_intidxs(
ignore_overlaps=False,
ignore_upstream=False,
ignore_downstream=False,
+ direction_col=None,
tie_breaking_col=None,
cols1=None,
cols2=None,
@@ -897,6 +985,15 @@ def _closest_intidxs(
"f(DataFrame) -> Series"
)
+ # Verify and construct the direction_arr (convert from pandas string column to bool array)
+ # TODO: should we add checks that it's valid "strand"?
+ direction_arr = None
+ if direction_col is None:
+ direction_arr = np.ones(len(df1_group), dtype=np.bool_)
+ else:
+ direction_arr = (df1_group[direction_col].values != "-") # both "+" and "." keep orientation by genomic coordinate
+
+ # Calculate closest intervals with arrops:
closest_idxs_group = arrops.closest_intervals(
df1_group[sk1].values,
df1_group[ek1].values,
@@ -907,6 +1004,7 @@ def _closest_intidxs(
ignore_overlaps=ignore_overlaps,
ignore_upstream=ignore_upstream,
ignore_downstream=ignore_downstream,
+ direction=direction_arr
)
# Convert local per-chromosome indices into the
@@ -934,6 +1032,7 @@ def closest(
ignore_overlaps=False,
ignore_upstream=False,
ignore_downstream=False,
+ direction_col=None,
tie_breaking_col=None,
return_input=True,
return_index=False,
@@ -946,6 +1045,9 @@ def closest(
"""
For every interval in dataframe `df1` find k closest genomic intervals in dataframe `df2`.
+ Currently, we are not taking the feature strands into account for filtering.
+ However, the strand can be used for definition of upstream/downstream of the feature (direction).
+
Note that, unless specified otherwise, overlapping intervals are considered as closest.
When multiple intervals are located at the same distance, the ones with the lowest index
in `df2` are returned.
@@ -957,20 +1059,22 @@ def closest(
If `df2` is None, find closest non-identical intervals within the same set.
k : int
- The number of closest intervals to report.
+ The number of the closest intervals to report.
ignore_overlaps : bool
- If True, return the closest non-overlapping interval.
+ If True, ignore overlapping intervals and return the closest non-overlapping interval.
ignore_upstream : bool
- If True, ignore intervals in `df2` that are upstream (relative to the
- reference strand) of intervals in `df1`. Currently, we are not taking
- the feature strands into account.
+ If True, ignore intervals in `df2` that are upstream of intervals in `df1`,
+ relative to the reference strand or the strand specified by direction_col.
ignore_downstream : bool
- If True, ignore intervals in `df2` that are downstream (relative to the
- reference strand) of intervals in `df1`. Currently, we are not taking
- the feature strands into account.
+ If True, ignore intervals in `df2` that are downstream of intervals in `df1`,
+ relative to the reference strand or the strand specified by direction_col.
+
+ direction_col : str
+ Name of direction column that will set upstream/downstream orientation for each feature.
+ The column should contain bioframe-compliant strand values ("+", "-", ".").
tie_breaking_col : str
A column in `df2` to use for breaking ties when multiple intervals
@@ -1005,6 +1109,17 @@ def closest(
df_closest : pandas.DataFrame
If no intervals found, returns none.
+ Notes
+ -----
+ By default, direction is defined by the reference genome: everything with
+ smaller coordinate is considered upstream, everything with larger coordinate
+ is considered downstream.
+
+ If ``direction_col`` is provided, upstream/downstream are relative to the
+ direction column in ``df1``, i.e. features marked "+" and "." strand will
+ define upstream and downstream as above, while features marked "-" have
+ upstream and downstream reversed: smaller coordinates are downstream and
+ larger coordinates are upstream.
"""
if k < 1:
@@ -1031,6 +1146,7 @@ def closest(
ignore_overlaps=ignore_overlaps,
ignore_upstream=ignore_upstream,
ignore_downstream=ignore_downstream,
+ direction_col=direction_col,
tie_breaking_col=tie_breaking_col,
cols1=cols1,
cols2=cols2,
=====================================
bioframe/sandbox/clients.py
=====================================
@@ -8,7 +8,7 @@ import socket
import base64
import glob
-from .fileops import read_table
+from ..io.fileops import read_table
class EncodeClient:
=====================================
docs/figs/bioframe_closest.pdf
=====================================
Binary files /dev/null and b/docs/figs/bioframe_closest.pdf differ
=====================================
docs/figs/closest0.png
=====================================
Binary files /dev/null and b/docs/figs/closest0.png differ
=====================================
docs/figs/closest1.png
=====================================
Binary files /dev/null and b/docs/figs/closest1.png differ
=====================================
docs/figs/closest2.png
=====================================
Binary files /dev/null and b/docs/figs/closest2.png differ
=====================================
docs/figs/closest3.png
=====================================
Binary files /dev/null and b/docs/figs/closest3.png differ
=====================================
docs/guide-definitions.rst
=====================================
@@ -35,7 +35,7 @@ View (i.e. a set of Genomic Regions):
- We define views separately from the scaffolds that make up a genome assembly, as a set of more constrained and ordered genomic regions are often useful for downstream analysis and visualization.
- An assembly is a special case of a view, where the individual regions correspond to the assembly’s entire scaffolds.
-Associating sets of genomic intervals with views
+Associating genomic intervals with views
- Similarly to how genomic intervals are associated with a scaffold, they can also be associated with a region from a view with an additional string, making a quadruple (chrom, start, end, view_region). This string must be *cataloged* in the view, i.e. it must match the name of a region in the view. Typically the interval would be contained in its associated view region, or, at the minimum, have a greater overlap with that region than other view regions.
- If each interval in a set is contained in their associated view region, the set is *contained* in the view.
- A set of intervals *covers* a view if each region in the view is contained by the union of its associated intervals. Conversely, if a set does not cover all of view regions, the interval set will have *gaps* relative to that view (stretches of bases not covered by an interval).
=====================================
docs/guide-intervalops.md
=====================================
@@ -219,6 +219,28 @@ Closest intervals within a single DataFrame can be found simply by passing a sin
bf.closest(df1, k=2)
```
+```{eval-rst}
+Closest intervals upstream of the features in df1 can be found by ignoring downstream and overlaps.
+Upstream/downstream direction is defined by genomic coordinates by default (smaller coordinate is upstream).
+```
+```{code-cell} ipython3
+bf.closest(df1, df2,
+ ignore_overlaps=True,
+ ignore_downstream=True)
+```
+
+```{eval-rst}
+If the features in df1 have direction (e.g., genes have transcription direction), then the definition of upstream/downstream
+direction can be changed to the direction of the features by `direction_col`:
+```
+```{code-cell} ipython3
+bf.closest(df1, df2,
+ ignore_overlaps=True,
+ ignore_downstream=True,
+ direction_col='strand')
+```
+
+
## Coverage & Count Overlaps
```{eval-rst}
For two sets of genomic features, it is often useful to calculate the number of basepairs covered and the number of overlapping intervals. While these are fairly straightforward to compute from the output of :func:`bioframe.overlap` with :func:`pandas.groupby` and column renaming, since these are very frequently used, they are provided as core bioframe functions..
=====================================
docs/guide-recipes.md
=====================================
@@ -52,6 +52,17 @@ Use closest after filtering by strand, and passing the `ignore_upsream=True` arg
bioframe.closest(df1.loc[df1['strand']=='+'], df2, ignore_upstream=True)
```
+For gener, the upstream/downstream direction might be defined by the direction of transcription.
+Use `direction_col='strand'` to set up the direction:
+```
+bioframe.closest(df1, df2, ignore_upstream=True, direction_col='strand')
+```
+
+## Drop non-autosomes from a bedframe?
+Use pandas DataFrame.isin(values):
+```
+df[ ~df.chrom.isin(['chrX','chrY'])]
+```
=====================================
docs/guide-specifications.rst
=====================================
@@ -0,0 +1,36 @@
+.. _Specifications:
+
+Specifications
+===========
+
+BedFrame (i.e. genomic intervals stored in a pandas dataframe):
+ - In a BedFrame, three required columns specify the set of genomic intervals (default column names = (‘chrom’, ‘start’, ‘end’)).
+ - Other reserved but not required column names: (‘strand’, ‘name’, ‘view_region’).
+
+ - entries in column ‘name’ are expected to be unique
+ - ‘view_region’ is expected to point to an associated region in a view with a matching name
+ - ‘strand’ is expected to be encoded with strings (‘+’, ‘-’, ‘.’).
+
+ - Additional columns are allowed: ‘zodiac_sign’, ‘soundcloud’, ‘twitter_name’, etc.
+ - Repeated intervals are allowed.
+ - The native pandas DataFrame index is not intended to be used as an immutable lookup table for genomic intervals in BedFrame. This is because many common genomic interval operations change the number of intervals stored in a BedFrame.
+ - Two useful sorting schemes for BedFrames are:
+
+ - scaffold-sorted: on (chrom, start, end), where chrom is sorted lexicographically.
+ - view-sorted: on (view_region, start, end) where view_region is sorted by order in the view.
+
+ - Null values are allowed, but only as pd.NA (using np.nan is discouraged as it results in unwanted type re-casting).
+ - Note if no ‘view_region’ is assigned to a genomic interval, then ‘chrom’ implicitly defines an associated region
+ - Note the BedFrame specification is a natural extension of the BED format ( https://samtools.github.io/hts-specs/BEDv1.pdf ) for pandas DataFrames.
+
+ViewFrames (a genomic view stored in a pandas dataframe)
+ - BedFrame where:
+
+ - intervals are non-overlapping
+ - “name” column is mandatory and contains a set of unique strings.
+
+ - Note that a ViewFrame can potentially be indexed by the name column to serve as a lookup table. This functionality is currently not implemented, because within the current Pandas implementation indexing by a column removes the column from the table.
+ - Note that views can be defined by:
+
+ - dictionary of string:ints (start=0 assumed) or string:tuples (start,end)
+ - pandas series of chromsizes (start=0, name=chrom)
=====================================
docs/index.rst
=====================================
@@ -19,12 +19,14 @@ bioframe
guide-performance.ipynb
guide-recipes.md
guide-definitions
+ guide-specifications
.. toctree::
:maxdepth: 1
:caption: Tutorials
tutorials/tutorial_assign_motifs_to_peaks.ipynb
+ tutorials/tutorial_assign_peaks_to_genes.ipynb
.. toctree::
:maxdepth: 3
=====================================
docs/tutorials/tutorial_assign_motifs_to_peaks.ipynb
=====================================
The diff for this file was not included because it is too large.
=====================================
docs/tutorials/tutorial_assign_peaks_to_genes.ipynb
=====================================
@@ -0,0 +1,627 @@
+{
+ "cells": [
+ {
+ "cell_type": "markdown",
+ "id": "57c80a2c",
+ "metadata": {},
+ "source": [
+ "# How to: assign ChIP-seq peaks to genes\n",
+ "\n",
+ "This tutorial demonstrates one way to assign CTCF ChIP-seq peaks to the nearest genes using bioframe."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 1,
+ "id": "ad9ab941",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "import bioframe\n",
+ "import numpy as np\n",
+ "import pandas as pd \n",
+ "import matplotlib.pyplot as plt"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 72,
+ "id": "562865cc",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "base_dir = '/tmp/bioframe_tutorial_data/'\n",
+ "assembly = 'hg38'"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "d3dae5c3",
+ "metadata": {},
+ "source": [
+ "## Load chromosome sizes\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 74,
+ "id": "6253803a",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "text/plain": [
+ "chr21 46709983\n",
+ "chr22 50818468\n",
+ "chrX 156040895\n",
+ "chrY 57227415\n",
+ "chrM 16569\n",
+ "Name: length, dtype: int64"
+ ]
+ },
+ "execution_count": 74,
+ "metadata": {},
+ "output_type": "execute_result"
+ }
+ ],
+ "source": [
+ "chromsizes = bioframe.fetch_chromsizes(assembly)\n",
+ "chromsizes.tail()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 78,
+ "id": "c74347d2",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "chromosomes = bioframe.make_viewframe(chromsizes)"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "eb8e2724",
+ "metadata": {},
+ "source": [
+ "## Load CTCF ChIP-seq peaks for HFF from ENCODE\n",
+ "\n",
+ "This approach makes use of the `narrowPeak` schema for bioframe.read_table . "
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 3,
+ "id": "48616968",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "text/html": [
+ "<div>\n",
+ "<style scoped>\n",
+ " .dataframe tbody tr th:only-of-type {\n",
+ " vertical-align: middle;\n",
+ " }\n",
+ "\n",
+ " .dataframe tbody tr th {\n",
+ " vertical-align: top;\n",
+ " }\n",
+ "\n",
+ " .dataframe thead th {\n",
+ " text-align: right;\n",
+ " }\n",
+ "</style>\n",
+ "<table border=\"1\" class=\"dataframe\">\n",
+ " <thead>\n",
+ " <tr style=\"text-align: right;\">\n",
+ " <th></th>\n",
+ " <th>chrom</th>\n",
+ " <th>start</th>\n",
+ " <th>end</th>\n",
+ " <th>name</th>\n",
+ " <th>score</th>\n",
+ " <th>strand</th>\n",
+ " <th>fc</th>\n",
+ " <th>-log10p</th>\n",
+ " <th>-log10q</th>\n",
+ " <th>relSummit</th>\n",
+ " </tr>\n",
+ " </thead>\n",
+ " <tbody>\n",
+ " <tr>\n",
+ " <th>0</th>\n",
+ " <td>chr19</td>\n",
+ " <td>48309541</td>\n",
+ " <td>48309911</td>\n",
+ " <td>.</td>\n",
+ " <td>1000</td>\n",
+ " <td>.</td>\n",
+ " <td>5.04924</td>\n",
+ " <td>-1.0</td>\n",
+ " <td>0.00438</td>\n",
+ " <td>185</td>\n",
+ " </tr>\n",
+ " <tr>\n",
+ " <th>1</th>\n",
+ " <td>chr4</td>\n",
+ " <td>130563716</td>\n",
+ " <td>130564086</td>\n",
+ " <td>.</td>\n",
+ " <td>993</td>\n",
+ " <td>.</td>\n",
+ " <td>5.05052</td>\n",
+ " <td>-1.0</td>\n",
+ " <td>0.00432</td>\n",
+ " <td>185</td>\n",
+ " </tr>\n",
+ " <tr>\n",
+ " <th>2</th>\n",
+ " <td>chr1</td>\n",
+ " <td>200622507</td>\n",
+ " <td>200622877</td>\n",
+ " <td>.</td>\n",
+ " <td>591</td>\n",
+ " <td>.</td>\n",
+ " <td>5.05489</td>\n",
+ " <td>-1.0</td>\n",
+ " <td>0.00400</td>\n",
+ " <td>185</td>\n",
+ " </tr>\n",
+ " <tr>\n",
+ " <th>3</th>\n",
+ " <td>chr5</td>\n",
+ " <td>112848447</td>\n",
+ " <td>112848817</td>\n",
+ " <td>.</td>\n",
+ " <td>869</td>\n",
+ " <td>.</td>\n",
+ " <td>5.05841</td>\n",
+ " <td>-1.0</td>\n",
+ " <td>0.00441</td>\n",
+ " <td>185</td>\n",
+ " </tr>\n",
+ " <tr>\n",
+ " <th>4</th>\n",
+ " <td>chr1</td>\n",
+ " <td>145960616</td>\n",
+ " <td>145960986</td>\n",
+ " <td>.</td>\n",
+ " <td>575</td>\n",
+ " <td>.</td>\n",
+ " <td>5.05955</td>\n",
+ " <td>-1.0</td>\n",
+ " <td>0.00439</td>\n",
+ " <td>185</td>\n",
+ " </tr>\n",
+ " </tbody>\n",
+ "</table>\n",
+ "</div>"
+ ],
+ "text/plain": [
+ " chrom start end name score strand fc -log10p -log10q \\\n",
+ "0 chr19 48309541 48309911 . 1000 . 5.04924 -1.0 0.00438 \n",
+ "1 chr4 130563716 130564086 . 993 . 5.05052 -1.0 0.00432 \n",
+ "2 chr1 200622507 200622877 . 591 . 5.05489 -1.0 0.00400 \n",
+ "3 chr5 112848447 112848817 . 869 . 5.05841 -1.0 0.00441 \n",
+ "4 chr1 145960616 145960986 . 575 . 5.05955 -1.0 0.00439 \n",
+ "\n",
+ " relSummit \n",
+ "0 185 \n",
+ "1 185 \n",
+ "2 185 \n",
+ "3 185 \n",
+ "4 185 "
+ ]
+ },
+ "execution_count": 3,
+ "metadata": {},
+ "output_type": "execute_result"
+ }
+ ],
+ "source": [
+ "ctcf_peaks = bioframe.read_table(\"https://www.encodeproject.org/files/ENCFF401MQL/@@download/ENCFF401MQL.bed.gz\", schema='narrowPeak')\n",
+ "ctcf_peaks.head()"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 83,
+ "id": "5a228b72",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# Filter for selected chromosomes:\n",
+ "ctcf_peaks = bioframe.overlap(ctcf_peaks, chromosomes).dropna(subset=['name_'])[ctcf_peaks.columns]"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "e39fca85",
+ "metadata": {},
+ "source": [
+ "## Get list of genes from UCSC\n",
+ "\n",
+ "UCSC genes are stored in .gtf format."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 4,
+ "id": "e75ffbb4",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "text/html": [
+ "<div>\n",
+ "<style scoped>\n",
+ " .dataframe tbody tr th:only-of-type {\n",
+ " vertical-align: middle;\n",
+ " }\n",
+ "\n",
+ " .dataframe tbody tr th {\n",
+ " vertical-align: top;\n",
+ " }\n",
+ "\n",
+ " .dataframe thead th {\n",
+ " text-align: right;\n",
+ " }\n",
+ "</style>\n",
+ "<table border=\"1\" class=\"dataframe\">\n",
+ " <thead>\n",
+ " <tr style=\"text-align: right;\">\n",
+ " <th></th>\n",
+ " <th>chrom</th>\n",
+ " <th>source</th>\n",
+ " <th>feature</th>\n",
+ " <th>start</th>\n",
+ " <th>end</th>\n",
+ " <th>score</th>\n",
+ " <th>strand</th>\n",
+ " <th>frame</th>\n",
+ " <th>attributes</th>\n",
+ " </tr>\n",
+ " </thead>\n",
+ " <tbody>\n",
+ " <tr>\n",
+ " <th>47</th>\n",
+ " <td>chr1</td>\n",
+ " <td>ensGene</td>\n",
+ " <td>CDS</td>\n",
+ " <td>69091</td>\n",
+ " <td>70005</td>\n",
+ " <td>.</td>\n",
+ " <td>+</td>\n",
+ " <td>0</td>\n",
+ " <td>gene_id \"ENSG00000186092\"; transcript_id \"ENST...</td>\n",
+ " </tr>\n",
+ " <tr>\n",
+ " <th>112</th>\n",
+ " <td>chr1</td>\n",
+ " <td>ensGene</td>\n",
+ " <td>CDS</td>\n",
+ " <td>182709</td>\n",
+ " <td>182746</td>\n",
+ " <td>.</td>\n",
+ " <td>+</td>\n",
+ " <td>0</td>\n",
+ " <td>gene_id \"ENSG00000279928\"; transcript_id \"ENST...</td>\n",
+ " </tr>\n",
+ " <tr>\n",
+ " <th>114</th>\n",
+ " <td>chr1</td>\n",
+ " <td>ensGene</td>\n",
+ " <td>CDS</td>\n",
+ " <td>183114</td>\n",
+ " <td>183240</td>\n",
+ " <td>.</td>\n",
+ " <td>+</td>\n",
+ " <td>1</td>\n",
+ " <td>gene_id \"ENSG00000279928\"; transcript_id \"ENST...</td>\n",
+ " </tr>\n",
+ " <tr>\n",
+ " <th>116</th>\n",
+ " <td>chr1</td>\n",
+ " <td>ensGene</td>\n",
+ " <td>CDS</td>\n",
+ " <td>183922</td>\n",
+ " <td>184155</td>\n",
+ " <td>.</td>\n",
+ " <td>+</td>\n",
+ " <td>0</td>\n",
+ " <td>gene_id \"ENSG00000279928\"; transcript_id \"ENST...</td>\n",
+ " </tr>\n",
+ " <tr>\n",
+ " <th>122</th>\n",
+ " <td>chr1</td>\n",
+ " <td>ensGene</td>\n",
+ " <td>CDS</td>\n",
+ " <td>185220</td>\n",
+ " <td>185350</td>\n",
+ " <td>.</td>\n",
+ " <td>-</td>\n",
+ " <td>2</td>\n",
+ " <td>gene_id \"ENSG00000279457\"; transcript_id \"ENST...</td>\n",
+ " </tr>\n",
+ " </tbody>\n",
+ "</table>\n",
+ "</div>"
+ ],
+ "text/plain": [
+ " chrom source feature start end score strand frame \\\n",
+ "47 chr1 ensGene CDS 69091 70005 . + 0 \n",
+ "112 chr1 ensGene CDS 182709 182746 . + 0 \n",
+ "114 chr1 ensGene CDS 183114 183240 . + 1 \n",
+ "116 chr1 ensGene CDS 183922 184155 . + 0 \n",
+ "122 chr1 ensGene CDS 185220 185350 . - 2 \n",
+ "\n",
+ " attributes \n",
+ "47 gene_id \"ENSG00000186092\"; transcript_id \"ENST... \n",
+ "112 gene_id \"ENSG00000279928\"; transcript_id \"ENST... \n",
+ "114 gene_id \"ENSG00000279928\"; transcript_id \"ENST... \n",
+ "116 gene_id \"ENSG00000279928\"; transcript_id \"ENST... \n",
+ "122 gene_id \"ENSG00000279457\"; transcript_id \"ENST... "
+ ]
+ },
+ "execution_count": 4,
+ "metadata": {},
+ "output_type": "execute_result"
+ }
+ ],
+ "source": [
+ "genes_url = 'https://hgdownload.cse.ucsc.edu/goldenpath/hg38/bigZips/genes/hg38.ensGene.gtf.gz'\n",
+ "genes = bioframe.read_table(genes_url, schema='gtf').query('feature==\"CDS\"')\n",
+ "\n",
+ "genes.head() \n",
+ "\n",
+ "## Note this functions to parse the attributes of the genes:\n",
+ "# import bioframe.sandbox.gtf_io\n",
+ "# genes_attr = bioframe.sandbox.gtf_io.parse_gtf_attributes(genes['attributes'])"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 84,
+ "id": "84b4226f",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# Filter for selected chromosomes:\n",
+ "genes = bioframe.overlap(genes, chromosomes).dropna(subset=['name_'])[genes.columns]"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "111ff194",
+ "metadata": {},
+ "source": [
+ "## Assign each peak to the gene\n",
+ "\n",
+ "\n",
+ "\n",
+ "\n",
+ "\n",
+ "Here, we want to assign each peak (feature) to a gene (input table)."
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 85,
+ "id": "4d78c70b",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "peaks_closest = bioframe.closest(genes, ctcf_peaks)"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 87,
+ "id": "b55e2e12",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "text/plain": [
+ "(0.0, 1000.0)"
+ ]
+ },
+ "execution_count": 87,
+ "metadata": {},
+ "output_type": "execute_result"
+ },
+ {
+ "data": {
+ "image/png": "iVBORw0KGgoAAAANSUhEUgAAAksAAAGdCAYAAAACMjetAAAAOXRFWHRTb2Z0d2FyZQBNYXRwbG90bGliIHZlcnNpb24zLjQuMywgaHR0cHM6Ly9tYXRwbG90bGliLm9yZy/MnkTPAAAACXBIWXMAAA9hAAAPYQGoP6dpAAAmi0lEQVR4nO3df1DU953H8RfyYwUOvicS2GyDFmcYo8W0FnOI8ao9Fb1KmE5vqg1ma6aemjP+2Kr1x6V3tZkLGNtqpuVq1MvEXDQluUns5VKPStocjSeKh9L6M7lOiaKyYtNl0YSCwc/9kfE7t2I+1bj8fj5m9o/97pvd73c/Ik+/7K4xxhgjAAAA3NSQ3t4BAACAvoxYAgAAsCCWAAAALIglAAAAC2IJAADAglgCAACwIJYAAAAsiCUAAACLuN7egd507do1XbhwQSkpKYqJient3QEAALfAGKPLly/L5/NpyJDuP+8zqGPpwoULysrK6u3dAAAAn0BjY6Puueeebn+cQR1LKSkpkj56slNTU3t5bwAAwK1obW1VVlaW+3O8uw3qWLr+q7fU1FRiCQCAfqanXkLDC7wBAAAsiCUAAAALYgkAAMCCWAIAALAglgAAACyIJQAAAAtiCQAAwIJYAgAAsCCWAAAALIglAAAAC2IJAADAglgCAACwIJYAAAAsiCUAAACLuN7egb4g9zs/1xBPknv93Y2ze3FvAABAX8KZJQAAAAtiCQAAwIJYAgAAsCCWAAAALIglAAAAC2IJAADAglgCAACwIJYAAAAsiCUAAAALYgkAAMCCWAIAALAglgAAACyIJQAAAAtiCQAAwIJYAgAAsCCWAAAALIglAAAAC2IJAADAglgCAACwIJYAAAAsiCUAAAALYgkAAMCCWAIAALAglgAAACyIJQAAAAtiCQAAwIJYAgAAsCCWAAAALIglAAAAC2IJAADAglgCAACwIJYAAAAsiCUAAACL246lX/3qV3rwwQfl8/kUExOjn/70pxG3G2O0YcMG+Xw+JSYmaurUqTpx4kTETHt7u5YtW6b09HQlJyeruLhY586di5gJhULy+/1yHEeO48jv96ulpSVi5uzZs3rwwQeVnJys9PR0LV++XB0dHbd7SAAAAB/rtmPp/fff12c/+1mVl5ff9PZNmzZp8+bNKi8v1+HDh+X1ejVjxgxdvnzZnQkEAtqzZ48qKiq0f/9+XblyRUVFRers7HRnSkpKVF9fr8rKSlVWVqq+vl5+v9+9vbOzU7Nnz9b777+v/fv3q6KiQq+88opWrVp1u4cEAADw8cwdkGT27NnjXr927Zrxer1m48aN7rY//vGPxnEc88wzzxhjjGlpaTHx8fGmoqLCnTl//rwZMmSIqaysNMYYc/LkSSPJHDx40J2pqakxkszp06eNMcbs3bvXDBkyxJw/f96d+clPfmI8Ho8Jh8O3tP/hcNhIMlmBl83Ita+7FwAA0Hdd//l9qz/v71RUX7PU0NCgYDCowsJCd5vH49GUKVN04MABSVJdXZ2uXr0aMePz+ZSbm+vO1NTUyHEc5efnuzMTJ06U4zgRM7m5ufL5fO7MzJkz1d7errq6upvuX3t7u1pbWyMuAAAANlGNpWAwKEnKzMyM2J6ZmeneFgwGlZCQoGHDhllnMjIyutx/RkZGxMyNjzNs2DAlJCS4MzcqKytzXwPlOI6ysrI+wVECAIDBpFveDRcTExNx3RjTZduNbpy52fwnmfn/1q9fr3A47F4aGxut+wQAABDVWPJ6vZLU5cxOc3OzexbI6/Wqo6NDoVDIOnPx4sUu93/p0qWImRsfJxQK6erVq13OOF3n8XiUmpoacQEAALCJaixlZ2fL6/WqqqrK3dbR0aHq6mpNmjRJkpSXl6f4+PiImaamJh0/ftydKSgoUDgcVm1trTtz6NAhhcPhiJnjx4+rqanJndm3b588Ho/y8vKieVgAAGAQi7vdL7hy5Yp++9vfutcbGhpUX1+vtLQ0jRgxQoFAQKWlpcrJyVFOTo5KS0uVlJSkkpISSZLjOFqwYIFWrVql4cOHKy0tTatXr9a4ceM0ffp0SdKYMWM0a9YsLVy4UNu2bZMkLVq0SEVFRRo9erQkqbCwUGPHjpXf79f3vvc9/eEPf9Dq1au1cOFCzhgBAICoue1Y+p//+R998YtfdK+vXLlSkjR//nzt3LlTa9asUVtbm5YsWaJQKKT8/Hzt27dPKSkp7tds2bJFcXFxmjNnjtra2jRt2jTt3LlTsbGx7szu3bu1fPly911zxcXFEZ/tFBsbq5/97GdasmSJHnjgASUmJqqkpETf//73b/9ZAAAA+BgxxhjT2zvRW1pbWz96V1zgZQ3xJLnb3904uxf3CgAA2Fz/+R0Oh3vkt0n833AAAAAWxBIAAIAFsQQAAGBBLAEAAFgQSwAAABbEEgAAgAWxBAAAYEEsAQAAWBBLAAAAFsQSAACABbEEAABgQSwBAABYEEsAAAAWxBIAAIAFsQQAAGBBLAEAAFgQSwAAABbEEgAAgAWxBAAAYEEsAQAAWBBLAAAAFsQSAACABbEEAABgQSwBAABYEEsAAAAWxBIAAIAFsQQAAGBBLAEAAFgQSwAAABbEEgAAgAWxBAAAYEEsAQAAWBBLAAAAFsQSAACABbEEAABgQSwBAABYEEsAAAAWxBIAAIAFsQQAAGBBLAEAAFgQSwAAABbEEgAAgAWxBAAAYEEsAQAAWBBLAAAAFsQSAACABbEEAABgQSwBAABYEEsAAAAWxBIAAIAFsQQAAGBBLAEAAFhEPZY+/PBDffvb31Z2drYSExM1atQoPfHEE7p27Zo7Y4zRhg0b5PP5lJiYqKlTp+rEiRMR99Pe3q5ly5YpPT1dycnJKi4u1rlz5yJmQqGQ/H6/HMeR4zjy+/1qaWmJ9iEBAIBBLOqx9NRTT+mZZ55ReXm5Tp06pU2bNul73/uefvSjH7kzmzZt0ubNm1VeXq7Dhw/L6/VqxowZunz5sjsTCAS0Z88eVVRUaP/+/bpy5YqKiorU2dnpzpSUlKi+vl6VlZWqrKxUfX29/H5/tA8JAAAMYjHGGBPNOywqKlJmZqaeffZZd9vf/M3fKCkpSS+88IKMMfL5fAoEAlq7dq2kj84iZWZm6qmnntLixYsVDod111136YUXXtDcuXMlSRcuXFBWVpb27t2rmTNn6tSpUxo7dqwOHjyo/Px8SdLBgwdVUFCg06dPa/To0X9yX1tbW+U4jrICL2uIJ8nd/u7G2dF8SgAAQBRd//kdDoeVmpra7Y8X9TNLkydP1i9+8Qu98847kqRf//rX2r9/v770pS9JkhoaGhQMBlVYWOh+jcfj0ZQpU3TgwAFJUl1dna5evRox4/P5lJub687U1NTIcRw3lCRp4sSJchzHnblRe3u7WltbIy4AAAA2cdG+w7Vr1yocDuvee+9VbGysOjs79eSTT+qhhx6SJAWDQUlSZmZmxNdlZmbqzJkz7kxCQoKGDRvWZeb61weDQWVkZHR5/IyMDHfmRmVlZfrud797ZwcIAAAGlaifWXrppZe0a9cuvfjiizpy5Iief/55ff/739fzzz8fMRcTExNx3RjTZduNbpy52bztftavX69wOOxeGhsbb/WwAADAIBX1M0vf+ta3tG7dOn3ta1+TJI0bN05nzpxRWVmZ5s+fL6/XK+mjM0N33323+3XNzc3u2Sav16uOjg6FQqGIs0vNzc2aNGmSO3Px4sUuj3/p0qUuZ62u83g88ng80TlQAAAwKET9zNIHH3ygIUMi7zY2Ntb96IDs7Gx5vV5VVVW5t3d0dKi6utoNoby8PMXHx0fMNDU16fjx4+5MQUGBwuGwamtr3ZlDhw4pHA67MwAAAHcq6meWHnzwQT355JMaMWKEPvOZz+jo0aPavHmzvvGNb0j66FdngUBApaWlysnJUU5OjkpLS5WUlKSSkhJJkuM4WrBggVatWqXhw4crLS1Nq1ev1rhx4zR9+nRJ0pgxYzRr1iwtXLhQ27ZtkyQtWrRIRUVFt/ROOAAAgFsR9Vj60Y9+pH/4h3/QkiVL1NzcLJ/Pp8WLF+sf//Ef3Zk1a9aora1NS5YsUSgUUn5+vvbt26eUlBR3ZsuWLYqLi9OcOXPU1tamadOmaefOnYqNjXVndu/ereXLl7vvmisuLlZ5eXm0DwkAAAxiUf+cpf6Ez1kCAKD/6fefswQAADCQEEsAAAAWxBIAAIAFsQQAAGBBLAEAAFgQSwAAABbEEgAAgAWxBAAAYEEsAQAAWBBLAAAAFsQSAACABbEEAABgQSwBAABYEEsAAAAWxBIAAIAFsQQAAGBBLAEAAFgQSwAAABbEEgAAgAWxBAAAYEEsAQAAWBBLAAAAFsQSAACABbEEAABgQSwBAABYEEsAAAAWxBIAAIAFsQQAAGBBLAEAAFgQSwAAABbEEgAAgAWxBAAAYEEsAQAAWBBLAAAAFsQSAACABbEEAABgQSwBAABYEEsAAAAWxBIAAIAFsQQAAGBBLAEAAFgQSwAAABbEEgAAgAWxBAAAYEEsAQAAWBBLAAAAFsQSAACABbEEAABgQSwBAABYEEsAAAAWxBIAAIAFsQQAAGDRLbF0/vx5Pfzwwxo+fLiSkpL0uc99TnV1de7txhht2LBBPp9PiYmJmjp1qk6cOBFxH+3t7Vq2bJnS09OVnJys4uJinTt3LmImFArJ7/fLcRw5jiO/36+WlpbuOCQAADBIRT2WQqGQHnjgAcXHx+s///M/dfLkSf3gBz/Qn//5n7szmzZt0ubNm1VeXq7Dhw/L6/VqxowZunz5sjsTCAS0Z88eVVRUaP/+/bpy5YqKiorU2dnpzpSUlKi+vl6VlZWqrKxUfX29/H5/tA8JAAAMYjHGGBPNO1y3bp3++7//W2+99dZNbzfGyOfzKRAIaO3atZI+OouUmZmpp556SosXL1Y4HNZdd92lF154QXPnzpUkXbhwQVlZWdq7d69mzpypU6dOaezYsTp48KDy8/MlSQcPHlRBQYFOnz6t0aNH/8l9bW1tleM4ygq8rCGeJHf7uxtn3+nTAAAAusn1n9/hcFipqand/nhRP7P02muvacKECfrqV7+qjIwMjR8/Xjt27HBvb2hoUDAYVGFhobvN4/FoypQpOnDggCSprq5OV69ejZjx+XzKzc11Z2pqauQ4jhtKkjRx4kQ5juPO3Ki9vV2tra0RFwAAAJuox9Lvfvc7bd26VTk5Ofr5z3+uRx99VMuXL9e//uu/SpKCwaAkKTMzM+LrMjMz3duCwaASEhI0bNgw60xGRkaXx8/IyHBnblRWVua+vslxHGVlZd3ZwQIAgAEv6rF07do1ff7zn1dpaanGjx+vxYsXa+HChdq6dWvEXExMTMR1Y0yXbTe6ceZm87b7Wb9+vcLhsHtpbGy81cMCAACDVNRj6e6779bYsWMjto0ZM0Znz56VJHm9XknqcvanubnZPdvk9XrV0dGhUChknbl48WKXx7906VKXs1bXeTwepaamRlwAAABsoh5LDzzwgN5+++2Ibe+8845GjhwpScrOzpbX61VVVZV7e0dHh6qrqzVp0iRJUl5enuLj4yNmmpqadPz4cXemoKBA4XBYtbW17syhQ4cUDofdGQAAgDsVF+07/OY3v6lJkyaptLRUc+bMUW1trbZv367t27dL+uhXZ4FAQKWlpcrJyVFOTo5KS0uVlJSkkpISSZLjOFqwYIFWrVql4cOHKy0tTatXr9a4ceM0ffp0SR+drZo1a5YWLlyobdu2SZIWLVqkoqKiW3onHAAAwK2Ieizdf//92rNnj9avX68nnnhC2dnZevrppzVv3jx3Zs2aNWpra9OSJUsUCoWUn5+vffv2KSUlxZ3ZsmWL4uLiNGfOHLW1tWnatGnauXOnYmNj3Zndu3dr+fLl7rvmiouLVV5eHu1DAgAAg1jUP2epP+FzlgAA6H/6/ecsAQAADCTEEgAAgAWxBAAAYEEsAQAAWBBLAAAAFsQSAACABbEEAABgQSwBAABYEEsAAAAWxBIAAIAFsQQAAGBBLAEAAFgQSwAAABbEEgAAgAWxBAAAYEEsAQAAWBBLAAAAFsQSAACABbEEAABgQSwBAABYEEsAAAAWxBIAAIAFsQQAAGBBLAEAAFgQSwAAABbEEgAAgAWxBAAAYEEsAQAAWBBLAAAAFsQSAACABbEEAABgQSwBAABYEEsAAAAWxBIAAIAFsQQAAGBBLAEAAFgQSwAAABbEEgAAgAWxBAAAYEEsAQAAWBBLAAAAFsQSAACABbEEAABgQSwBAABYEEsAAAAWxBIAAIAFsQQAAGBBLAEAAFgQSwAAABbEEgAAgAWxBAAAYNHtsVRWVqaYmBgFAgF3mzFGGzZskM/nU2JioqZOnaoTJ05EfF17e7uWLVum9PR0JScnq7i4WOfOnYuYCYVC8vv9chxHjuPI7/erpaWluw8JAAAMIt0aS4cPH9b27dt13333RWzftGmTNm/erPLych0+fFher1czZszQ5cuX3ZlAIKA9e/aooqJC+/fv15UrV1RUVKTOzk53pqSkRPX19aqsrFRlZaXq6+vl9/u785AAAMAg022xdOXKFc2bN087duzQsGHD3O3GGD399NN6/PHH9ZWvfEW5ubl6/vnn9cEHH+jFF1+UJIXDYT377LP6wQ9+oOnTp2v8+PHatWuXjh07pjfeeEOSdOrUKVVWVupf/uVfVFBQoIKCAu3YsUOvv/663n777e46LAAAMMh0Wyw99thjmj17tqZPnx6xvaGhQcFgUIWFhe42j8ejKVOm6MCBA5Kkuro6Xb16NWLG5/MpNzfXnampqZHjOMrPz3dnJk6cKMdx3BkAAIA7Fdcdd1pRUaEjR47o8OHDXW4LBoOSpMzMzIjtmZmZOnPmjDuTkJAQcUbq+sz1rw8Gg8rIyOhy/xkZGe7Mjdrb29Xe3u5eb21tvY2jAgAAg1HUzyw1NjZqxYoV2rVrl4YOHfqxczExMRHXjTFdtt3oxpmbzdvup6yszH0xuOM4ysrKsj4eAABA1GOprq5Ozc3NysvLU1xcnOLi4lRdXa0f/vCHiouLc88o3Xj2p7m52b3N6/Wqo6NDoVDIOnPx4sUuj3/p0qUuZ62uW79+vcLhsHtpbGy84+MFAAADW9Rjadq0aTp27Jjq6+vdy4QJEzRv3jzV19dr1KhR8nq9qqqqcr+mo6ND1dXVmjRpkiQpLy9P8fHxETNNTU06fvy4O1NQUKBwOKza2lp35tChQwqHw+7MjTwej1JTUyMuAAAANlF/zVJKSopyc3MjtiUnJ2v48OHu9kAgoNLSUuXk5CgnJ0elpaVKSkpSSUmJJMlxHC1YsECrVq3S8OHDlZaWptWrV2vcuHHuC8bHjBmjWbNmaeHChdq2bZskadGiRSoqKtLo0aOjfVgAAGCQ6pYXeP8pa9asUVtbm5YsWaJQKKT8/Hzt27dPKSkp7syWLVsUFxenOXPmqK2tTdOmTdPOnTsVGxvrzuzevVvLly933zVXXFys8vLyHj8eAAAwcMUYY0xv70RvaW1t/eiF3oGXNcST5G5/d+PsXtwrAABgc/3ndzgc7pGX1PB/wwEAAFgQSwAAABbEEgAAgAWxBAAAYEEsAQAAWBBLAAAAFsQSAACABbEEAABgQSwBAABYEEsAAAAWxBIAAIAFsQQAAGBBLAEAAFgQSwAAABbEEgAAgAWxBAAAYEEsAQAAWBBLAAAAFsQSAACABbEEAABgQSwBAABYEEsAAAAWxBIAAIAFsQQAAGBBLAEAAFgQSwAAABbEEgAAgAWxBAAAYEEsAQAAWBBLAAAAFsQSAACABbEEAABgQSwBAABYEEsAAAAWxBIAAIAFsQQAAGBBLAEAAFgQSwAAABbEEgAAgAWxBAAAYEEsAQAAWBBLAAAAFsQSAACABbEEAABgQSwBAABYEEsAAAAWxBIAAIAFsQQAAGBBLAEAAFgQSwAAABbEEgAAgAWxBAAAYBH1WCorK9P999+vlJQUZWRk6Mtf/rLefvvtiBljjDZs2CCfz6fExERNnTpVJ06ciJhpb2/XsmXLlJ6eruTkZBUXF+vcuXMRM6FQSH6/X47jyHEc+f1+tbS0RPuQAADAIBb1WKqurtZjjz2mgwcPqqqqSh9++KEKCwv1/vvvuzObNm3S5s2bVV5ersOHD8vr9WrGjBm6fPmyOxMIBLRnzx5VVFRo//79unLlioqKitTZ2enOlJSUqL6+XpWVlaqsrFR9fb38fn+0DwkAAAxiMcYY050PcOnSJWVkZKi6ulpf+MIXZIyRz+dTIBDQ2rVrJX10FikzM1NPPfWUFi9erHA4rLvuuksvvPCC5s6dK0m6cOGCsrKytHfvXs2cOVOnTp3S2LFjdfDgQeXn50uSDh48qIKCAp0+fVqjR4/+k/vW2toqx3GUFXhZQzxJ7vZ3N87uhmcCAABEw/Wf3+FwWKmpqd3+eN3+mqVwOCxJSktLkyQ1NDQoGAyqsLDQnfF4PJoyZYoOHDggSaqrq9PVq1cjZnw+n3Jzc92ZmpoaOY7jhpIkTZw4UY7juDM3am9vV2tra8QFAADApltjyRijlStXavLkycrNzZUkBYNBSVJmZmbEbGZmpntbMBhUQkKChg0bZp3JyMjo8pgZGRnuzI3Kysrc1zc5jqOsrKw7O0AAADDgdWssLV26VL/5zW/0k5/8pMttMTExEdeNMV223ejGmZvN2+5n/fr1CofD7qWxsfFWDgMAAAxi3RZLy5Yt02uvvaY333xT99xzj7vd6/VKUpezP83Nze7ZJq/Xq46ODoVCIevMxYsXuzzupUuXupy1us7j8Sg1NTXiAgAAYBP1WDLGaOnSpXr11Vf1y1/+UtnZ2RG3Z2dny+v1qqqqyt3W0dGh6upqTZo0SZKUl5en+Pj4iJmmpiYdP37cnSkoKFA4HFZtba07c+jQIYXDYXcGAADgTsVF+w4fe+wxvfjii/r3f/93paSkuGeQHMdRYmKiYmJiFAgEVFpaqpycHOXk5Ki0tFRJSUkqKSlxZxcsWKBVq1Zp+PDhSktL0+rVqzVu3DhNnz5dkjRmzBjNmjVLCxcu1LZt2yRJixYtUlFR0S29Ew4AAOBWRD2Wtm7dKkmaOnVqxPbnnntOjzzyiCRpzZo1amtr05IlSxQKhZSfn699+/YpJSXFnd+yZYvi4uI0Z84ctbW1adq0adq5c6diY2Pdmd27d2v58uXuu+aKi4tVXl4e7UMCAACDWLd/zlJfxucsAQDQ/wy4z1kCAADoz4glAAAAC2IJAADAglgCAACwIJYAAAAsiCUAAAALYgkAAMCCWAIAALAglgAAACyIJQAAAAtiCQAAwIJYAgAAsCCWAAAALIglAAAAC2IJAADAglgCAACwIJYAAAAsiCUAAAALYgkAAMCCWAIAALAglgAAACyIJQAAAAtiCQAAwIJYAgAAsCCWAAAALIglAAAAC2IJAADAglgCAACwIJYAAAAsiCUAAAALYgkAAMCCWAIAALAglgAAACyIJQAAAAtiCQAAwIJYAgAAsCCWAAAALIglAAAAi7je3oH+4tPrftZl27sbZ3/iuTt5DAAA0HM4swQAAGDBmaU+rifOVN1s7pM+Ju4MZxeBvo3v0cGJWOoBtxIjd3p/3f3N2hOP2Zf+EupL+wIMBAP5e2ogHxs+QizdRLTjpi+J5rHdyX31xF8k/AV2c7fyvPDcgT8D6An95c8ZsXQHBnJUDSa3uo63OtdbL/zvy38e+8tfiP9fb+0zbxLBnRgIfwb64jEQSwNEX/5BeTN3sr+99bV96TEGu2i/O7UnflV+M3fyA6CvnCWOtsH2K/+b6cv/QOpLf1Z6ErEE4Jb+AuzrQRHtM4TR/tre0t1R1Vs/xHsiSO9EX/qzcuO+RPt7ajCIMcaY3t6J3tLa2irHcZQVeFlDPEm9vTsAbqKv/IsaGCj64/fUjft8/ed3OBxWampqtz8+Z5YAABhE+noY9UXEEoA+jb/YAfQ2PsEbAADAglgCAACwIJYAAAAs+n0s/fjHP1Z2draGDh2qvLw8vfXWW729SwAAYADp17H00ksvKRAI6PHHH9fRo0f1l3/5l/rrv/5rnT17trd3DQAADBD9OpY2b96sBQsW6G//9m81ZswYPf3008rKytLWrVt7e9cAAMAA0W8/OqCjo0N1dXVat25dxPbCwkIdOHDgpl/T3t6u9vZ293o4HJYkXWv/oPt2FAAA3JHW1tabXu+pz9Xut7H0+9//Xp2dncrMzIzYnpmZqWAweNOvKSsr03e/+90u289vfaQ7dhEAAESB8/TNt7/33ntyHKfbH7/fxtJ1MTExEdeNMV22Xbd+/XqtXLnSvd7S0qKRI0fq7NmzPfJk4+O1trYqKytLjY2NPfLR9fh4rEXfwnr0HaxF3xEOhzVixAilpaX1yOP121hKT09XbGxsl7NIzc3NXc42XefxeOTxeLpsdxyHP/h9RGpqKmvRR7AWfQvr0XewFn3HkCE989LrfvsC74SEBOXl5amqqipie1VVlSZNmtRLewUAAAaafntmSZJWrlwpv9+vCRMmqKCgQNu3b9fZs2f16KOP9vauAQCAAaJfx9LcuXP13nvv6YknnlBTU5Nyc3O1d+9ejRw58pa+3uPx6Dvf+c5NfzWHnsVa9B2sRd/CevQdrEXf0dNrEWN66n13AAAA/VC/fc0SAABATyCWAAAALIglAAAAC2IJAADAYlDH0o9//GNlZ2dr6NChysvL01tvvdXbuzSglJWV6f7771dKSooyMjL05S9/WW+//XbEjDFGGzZskM/nU2JioqZOnaoTJ05EzLS3t2vZsmVKT09XcnKyiouLde7cuZ48lAGnrKxMMTExCgQC7jbWouecP39eDz/8sIYPH66kpCR97nOfU11dnXs7a9FzPvzwQ337299Wdna2EhMTNWrUKD3xxBO6du2aO8N6dI9f/epXevDBB+Xz+RQTE6Of/vSnEbdH63kPhULy+/1yHEeO48jv96ulpeX2dtYMUhUVFSY+Pt7s2LHDnDx50qxYscIkJyebM2fO9PauDRgzZ840zz33nDl+/Lipr683s2fPNiNGjDBXrlxxZzZu3GhSUlLMK6+8Yo4dO2bmzp1r7r77btPa2urOPProo+ZTn/qUqaqqMkeOHDFf/OIXzWc/+1nz4Ycf9sZh9Xu1tbXm05/+tLnvvvvMihUr3O2sRc/4wx/+YEaOHGkeeeQRc+jQIdPQ0GDeeOMN89vf/tadYS16zj/90z+Z4cOHm9dff900NDSYf/u3fzN/9md/Zp5++ml3hvXoHnv37jWPP/64eeWVV4wks2fPnojbo/W8z5o1y+Tm5poDBw6YAwcOmNzcXFNUVHRb+zpoY+kv/uIvzKOPPhqx7d577zXr1q3rpT0a+Jqbm40kU11dbYwx5tq1a8br9ZqNGze6M3/84x+N4zjmmWeeMcYY09LSYuLj401FRYU7c/78eTNkyBBTWVnZswcwAFy+fNnk5OSYqqoqM2XKFDeWWIues3btWjN58uSPvZ216FmzZ8823/jGNyK2feUrXzEPP/ywMYb16Ck3xlK0nveTJ08aSebgwYPuTE1NjZFkTp8+fcv7Nyh/DdfR0aG6ujoVFhZGbC8sLNSBAwd6aa8GvnA4LEnuf3zY0NCgYDAYsQ4ej0dTpkxx16Gurk5Xr16NmPH5fMrNzWWtPoHHHntMs2fP1vTp0yO2sxY957XXXtOECRP01a9+VRkZGRo/frx27Njh3s5a9KzJkyfrF7/4hd555x1J0q9//Wvt379fX/rSlySxHr0lWs97TU2NHMdRfn6+OzNx4kQ5jnNba9OvP8H7k/r973+vzs7OLv/hbmZmZpf/mBfRYYzRypUrNXnyZOXm5kqS+1zfbB3OnDnjziQkJGjYsGFdZlir21NRUaEjR47o8OHDXW5jLXrO7373O23dulUrV67U3//936u2tlbLly+Xx+PR17/+ddaih61du1bhcFj33nuvYmNj1dnZqSeffFIPPfSQJL43eku0nvdgMKiMjIwu95+RkXFbazMoY+m6mJiYiOvGmC7bEB1Lly7Vb37zG+3fv7/LbZ9kHVir29PY2KgVK1Zo3759Gjp06MfOsRbd79q1a5owYYJKS0slSePHj9eJEye0detWff3rX3fnWIue8dJLL2nXrl168cUX9ZnPfEb19fUKBALy+XyaP3++O8d69I5oPO83m7/dtRmUv4ZLT09XbGxsl6psbm7uUrG4c8uWLdNrr72mN998U/fcc4+73ev1SpJ1Hbxerzo6OhQKhT52Bn9aXV2dmpublZeXp7i4OMXFxam6ulo//OEPFRcX5z6XrEX3u/vuuzV27NiIbWPGjNHZs2cl8X3R0771rW9p3bp1+trXvqZx48bJ7/frm9/8psrKyiSxHr0lWs+71+vVxYsXu9z/pUuXbmttBmUsJSQkKC8vT1VVVRHbq6qqNGnSpF7aq4HHGKOlS5fq1Vdf1S9/+UtlZ2dH3J6dnS2v1xuxDh0dHaqurnbXIS8vT/Hx8REzTU1NOn78OGt1G6ZNm6Zjx46pvr7evUyYMEHz5s1TfX29Ro0axVr0kAceeKDLR2i888477n8AzvdFz/rggw80ZEjkj8LY2Fj3owNYj94Rree9oKBA4XBYtbW17syhQ4cUDodvb21u/bXqA8v1jw549tlnzcmTJ00gEDDJycnm3Xff7e1dGzD+7u/+zjiOY/7rv/7LNDU1uZcPPvjAndm4caNxHMe8+uqr5tixY+ahhx666VtD77nnHvPGG2+YI0eOmL/6q7/iLblR8P/fDWcMa9FTamtrTVxcnHnyySfN//7v/5rdu3ebpKQks2vXLneGteg58+fPN5/61Kfcjw549dVXTXp6ulmzZo07w3p0j8uXL5ujR4+ao0ePGklm8+bN5ujRo+5H+ETreZ81a5a57777TE1NjampqTHjxo3jowNuxz//8z+bkSNHmoSEBPP5z3/efUs7okPSTS/PPfecO3Pt2jXzne98x3i9XuPxeMwXvvAFc+zYsYj7aWtrM0uXLjVpaWkmMTHRFBUVmbNnz/bw0Qw8N8YSa9Fz/uM//sPk5uYaj8dj7r33XrN9+/aI21mLntPa2mpWrFhhRowYYYYOHWpGjRplHn/8cdPe3u7OsB7d480337zpz4j58+cbY6L3vL/33ntm3rx5JiUlxaSkpJh58+aZUCh0W/saY4wxn+AMGQAAwKAwKF+zBAAAcKuIJQAAAAtiCQAAwIJYAgAAsCCWAAAALIglAAAAC2IJAADAglgCAACwIJYAAAAsiCUAAAALYgkAAMCCWAIAALD4Pxjpf0OS2UyVAAAAAElFTkSuQmCC\n",
+ "text/plain": [
+ "<Figure size 640x480 with 1 Axes>"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ }
+ ],
+ "source": [
+ "# Plot the distribution of distances from peaks to genes: \n",
+ "plt.hist( peaks_closest['distance'], np.arange(0, 1e3, 10));\n",
+ "plt.xlim( [0, 1e3] )"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "8cec3987",
+ "metadata": {},
+ "source": [
+ "## Ignore upstream/downstream peaks from genes (strand-indifferent version)\n",
+ "\n",
+ "Sometimes you may want to ignore all the CTCFs upstream from the genes. \n",
+ "\n",
+ "By default, `bioframe.overlap` does not know the orintation of the genes, and thus assumes that the upstream/downstream is defined by the genomic coordinate (upstream is the direction towards the smaller coordinate):\n",
+ "\n",
+ "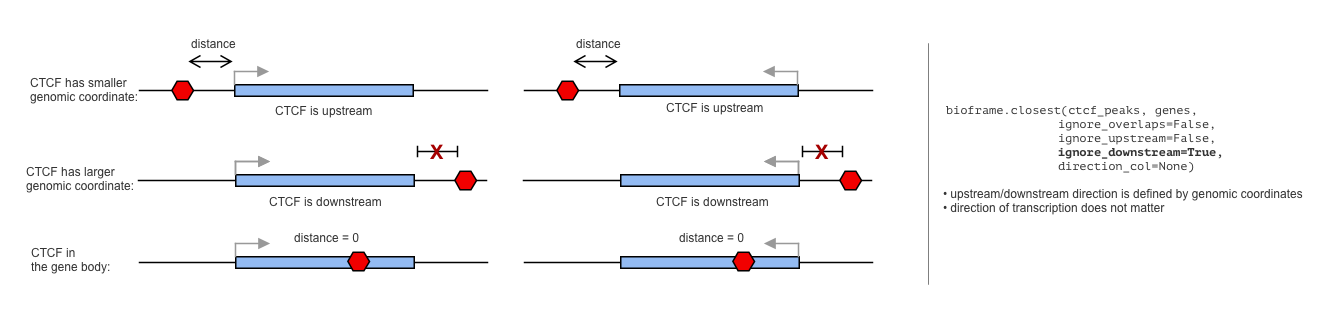\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 88,
+ "id": "e99e5213",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "peaks_closest_upstream_nodir = bioframe.closest(genes, ctcf_peaks, \n",
+ " ignore_overlaps=False,\n",
+ " ignore_upstream=False,\n",
+ " ignore_downstream=True,\n",
+ " direction_col=None)\n",
+ "\n",
+ "peaks_closest_downstream_nodir = bioframe.closest(genes, ctcf_peaks, \n",
+ " ignore_overlaps=False,\n",
+ " ignore_upstream=True,\n",
+ " ignore_downstream=False,\n",
+ " direction_col=None)"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "2fa693c9",
+ "metadata": {},
+ "source": [
+ "Note that distribution did not change much, and upstream and downstream distances are very similar:"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 92,
+ "id": "aa438234",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "text/plain": [
+ "<matplotlib.legend.Legend at 0x7fd148d1bfd0>"
+ ]
+ },
+ "execution_count": 92,
+ "metadata": {},
+ "output_type": "execute_result"
+ },
+ {
+ "data": {
+ "image/png": "iVBORw0KGgoAAAANSUhEUgAAAlAAAAGdCAYAAADdfE2yAAAAOXRFWHRTb2Z0d2FyZQBNYXRwbG90bGliIHZlcnNpb24zLjQuMywgaHR0cHM6Ly9tYXRwbG90bGliLm9yZy/MnkTPAAAACXBIWXMAAA9hAAAPYQGoP6dpAAA6YElEQVR4nO3de1RVdf7/8deRyxEQjlyE45nQ6BuZhqZiKeak5jVFai5pYaiTozamxqhp/ppmtJkkzbSVTqbVaJM2zDRl+W0cRmwSdcRLKI23MhsSSxArPIgiIOzfH37dqyOMsRM4oM/HWmet9me/z96fvTd2XuuzbzbDMAwBAACgzlp4uwMAAADNDQEKAADAIgIUAACARQQoAAAAiwhQAAAAFhGgAAAALCJAAQAAWESAAgAAsMjX2x3wpurqah0/flzBwcGy2Wze7g4AAKgDwzB0+vRpuVwutWjhnbGgazpAHT9+XNHR0d7uBgAA+B6OHTum6667zivrvqYDVHBwsKQLByAkJMTLvQEAAHVRUlKi6Oho83fcG67pAHXxtF1ISAgBCgCAZsabl99wETkAAIBFBCgAAACLCFAAAAAWXdPXQAEAvM8wDJ0/f15VVVXe7gqaCB8fH/n6+jbpRwwRoAAAXlNRUaGCggKdPXvW211BExMYGKi2bdvK39/f212pFQEKAOAV1dXVysvLk4+Pj1wul/z9/Zv0iAMah2EYqqio0MmTJ5WXl6fY2FivPSzzcghQAACvqKioUHV1taKjoxUYGOjt7qAJCQgIkJ+fn44ePaqKigq1bNnS212qoelFOgDANaUpji7A+5r630XT7h0AAEATRIACAACwiGugAABNypLMw426vl8OuqlR1/ddbDab1q1bp3vvvdfbXcFlMAIFAEAzU1lZ6e0uXPMIUAAAWHD99dfr+eef92jr2rWr5s6dK+nCCNLy5ct19913KyAgQDExMXrzzTfN2oqKCk2ZMkVt27ZVy5Ytdf311ystLc1ctiT96Ec/ks1mM6fnzp2rrl276g9/+INuuOEG2e12GYYht9utiRMnKjIyUiEhIbrrrrv00Ucfmev67LPPdM899ygqKkqtWrXSbbfdpk2bNtXYnt/97ncaM2aMWrVqpfbt2+vdd9/VyZMndc8996hVq1bq3LmzPvzww/rdkc0cAQoAgHr25JNP6ic/+Yk++ugjPfjgg3rggQd06NAhSdILL7yg9evX6y9/+Ys++eQTrVmzxgxKu3fvliStWrVKBQUF5rQkHTlyRH/5y1/01ltvKTc3V5I0fPhwFRYWasOGDcrJyVH37t01YMAAffPNN5Kk0tJSDRs2TJs2bdLevXs1ZMgQjRgxQvn5+R79XbJkie644w7t3btXw4cPV0pKisaMGaMHH3xQe/bs0Y033qgxY8bIMIwG3nPNB9dASfr9P4+oZVArc7qpnQ8HADQv9913n37+859Lkn77298qMzNTS5cu1Ysvvqj8/HzFxsaqT58+stlsat++vfm9Nm3aSJJat24tp9PpscyKigq9/vrrZs0///lP7du3T0VFRbLb7ZKkRYsW6Z133tFf//pXTZw4UbfeeqtuvfVWcxm/+93vtG7dOq1fv15Tpkwx24cNG6ZJkyZJkn79619r+fLluu2223TfffdJkmbPnq2EhASdOHGiRr+uVQQoSbd9sUpBAfZvtSzyWl8AAM1fQkJCjemLo0bjxo3ToEGD1KFDBw0dOlSJiYkaPHjwdy6zffv2ZniSpJycHJWWlio8PNyjrqysTJ999pkk6cyZM5o3b57ee+89HT9+XOfPn1dZWVmNEaguXbqY/x0VFSVJ6ty5c422oqIiAtT/IUABAGBBixYtapzKqstF3RdfU9O9e3fl5eXp73//uzZt2qSRI0dq4MCB+utf/3rZ7wcFBXlMV1dXq23bttq8eXON2tatW0uSHnvsMf3jH//QokWLdOONNyogIEA//elPVVFR4VHv5+dXo5+1tVVXV3/ndl4rCFAAAFjQpk0bFRQUmNMlJSXKy8vzqNmxY4fGjBnjMd2tWzdzOiQkRKNGjdKoUaP005/+VEOHDtU333yjsLAw+fn5qaqq6jv70b17dxUWFsrX19e8hupSW7du1bhx4/SjH/1I0oVroj7//HMLW4v/hgAFAIAFd911l1avXq0RI0YoNDRUTz75pHx8fDxq3nzzTfXo0UN9+vTR2rVrtWvXLr366quSLlyw3bZtW3Xt2lUtWrTQm2++KafTaY4aXX/99Xr//fd1xx13yG63KzQ0tNZ+DBw4UAkJCbr33nu1YMECdejQQcePH9eGDRt07733qkePHrrxxhv19ttva8SIEbLZbHryyScZRaon3IUHAIAFc+bM0Z133qnExEQNGzZM9957r/7nf/7Ho2bevHlKT09Xly5d9Nprr2nt2rXq1KmTJKlVq1ZasGCBevToodtuu02ff/65NmzYYL777bnnnlNmZqaio6M9Rq0uZbPZtGHDBt1555166KGHdNNNN+n+++/X559/bl6ztGTJEoWGhqp3794aMWKEhgwZou7duzfQnrm22Ixr+J7EkpISORwOZS6d4nERecJ4LiIHgIZ27tw55eXlKSYmRi1btvR2d+oNTxKvH5f7+7j4++12uxUSEuKV/jECBQAAYBEBCgAAwCIuIgcAoB5dw1fGXFMYgQIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAKgH/fr1U2pqqre7gUbCc6AAAE3LB2mNu77+cxp3fY3s888/V0xMjPbu3auuXbt6uztXDUagAACAKioqvN2FZoUABQCARWfOnNGYMWPUqlUrtW3bVs8995zH/OLiYo0ZM0ahoaEKDAzU3XffrU8//VTShSeVt2nTRm+99ZZZ37VrV0VGRprT2dnZ8vPzU2lpqaQLLyh+5ZVX9KMf/UiBgYGKjY3V+vXrPdY3evRotWnTRgEBAYqNjdWqVaskSTExMZKkbt26yWazqV+/fpKkcePG6d5771VaWppcLpduuukmSdKXX36pUaNGKTQ0VOHh4brnnnv0+eefm+vavXu3Bg0apIiICDkcDvXt21d79uzx2H6bzaYVK1YoMTFRgYGB6tixo7Kzs3XkyBH169dPQUFBSkhI0GeffXYlh8GrCFAAAFj02GOP6YMPPtC6deu0ceNGbd68WTk5Oeb8cePG6cMPP9T69euVnZ0twzA0bNgwVVZWymaz6c4779TmzZslXQg/Bw8eVGVlpQ4ePChJ2rx5s+Lj49WqVStzmfPmzdPIkSP173//W8OGDdPo0aP1zTffSJKefPJJHTx4UH//+9916NAhLV++XBEREZKkXbt2SZI2bdqkgoICvf322+Yy33//fR06dEiZmZl67733dPbsWfXv31+tWrXSli1btG3bNrVq1UpDhw41R6hOnz6tsWPHauvWrdqxY4diY2M1bNgwnT592mMf/fa3v9WYMWOUm5urm2++WcnJyZo0aZLmzJmjDz/8UJI0ZcqU+jwsjYproAAAsKC0tFSvvvqq/vjHP2rQoEGSpNdee03XXXedJOnTTz/V+vXr9a9//Uu9e/eWJK1du1bR0dF65513dN9996lfv35auXKlJGnLli269dZb1a5dO23evFmdOnXS5s2bzZGii8aNG6cHHnhAkjR//nwtXbpUu3bt0tChQ5Wfn69u3bqpR48ekqTrr7/e/F6bNm0kSeHh4XI6nR7LDAoK0iuvvCJ/f39J0h/+8Ae1aNFCr7zyimw2myRp1apVat26tTZv3qzBgwfrrrvu8ljGihUrFBoaqqysLCUmJprtP/vZzzRy5EhJ0uzZs5WQkKAnn3xSQ4YMkSQ9+uij+tnPfmZ19zcZjEABAGDBZ599poqKCiUkJJhtYWFh6tChgyTp0KFD8vX1Vc+ePc354eHh6tChgw4dOiTpwh17Bw4c0FdffaWsrCz169dP/fr1U1ZWls6fP6/t27erb9++Huvt0qWL+d9BQUEKDg5WUVGRJOkXv/iF0tPT1bVrV82aNUvbt2+v07Z07tzZDE+SlJOToyNHjig4OFitWrVSq1atFBYWpnPnzpmn24qKivTwww/rpptuksPhkMPhUGlpqfLz8/9rf6Oiosz1fbvt3LlzKikpqVNfmxpGoAAAsMAwjO813zAMc1QnLi5O4eHhysrKUlZWlp566ilFR0fr6aef1u7du1VWVqY+ffp4fN/Pz89j2mazqbq6WpJ099136+jRo/rb3/6mTZs2acCAAXrkkUe0aNGiy/Y1KCjIY7q6ulrx8fFau3ZtjdqLI1njxo3TyZMn9fzzz6t9+/ay2+1KSEiocRH6t/t7cbtra7u4Dc0NI1AAAFhw4403ys/PTzt27DDbiouLdfjwYUlSp06ddP78ee3cudOc//XXX+vw4cPq2LGjJJnXQb377rvav3+/fvjDH6pz586qrKzUSy+9pO7duys4ONhSv9q0aaNx48ZpzZo1ev75581ThBdHmKqqqr5zGd27d9enn36qyMhI3XjjjR4fh8MhSdq6daumTZumYcOG6ZZbbpHdbtdXX31lqa9XAwIUAAAWtGrVSuPHj9djjz2m999/X/v379e4cePUosWFn9TY2Fjdc889mjBhgrZt26aPPvpIDz74oH7wgx/onnvuMZfTr18/vfHGG+rSpYtCQkLMULV27doa1z99l1//+td69913deTIER04cEDvvfeeGdYiIyMVEBCgjIwMnThxQm63+78uZ/To0YqIiNA999yjrVu3Ki8vT1lZWXr00Uf1xRdfSLoQIF9//XUdOnRIO3fu1OjRoxUQEGBxLzZ/BCgAACx69tlndeeddyopKUkDBw5Unz59FB8fb85ftWqV4uPjlZiYqISEBBmGoQ0bNnicwurfv7+qqqo8wlLfvn1VVVVV4/qn7+Lv7685c+aoS5cuuvPOO+Xj46P09HRJkq+vr1544QWtWLFCLpfLI8RdKjAwUFu2bFG7du304x//WB07dtRDDz2ksrIyhYSESLpwoXlxcbG6deumlJQUTZs2zeMRDNcKm/FdJ3OvYiUlJXI4HMpcOkVBAXazPWH85c8ZAwCu3Llz55SXl6eYmBi1bNnS291BE3O5v4+Lv99ut9sMdo2NESgAAACLCFAAAAAWWQ5QW7Zs0YgRI+RyuWSz2fTOO++Y8yorKzV79mx17txZQUFBcrlcGjNmjI4fP+6xjPLyck2dOlUREREKCgpSUlKSeXHaRcXFxUpJSTGfMZGSkqJTp0551OTn52vEiBEKCgpSRESEpk2bxrt8AABAg7McoM6cOaNbb71Vy5YtqzHv7Nmz2rNnj5588knt2bNHb7/9tg4fPqykpCSPutTUVK1bt07p6enatm2bSktLlZiY6HGLZXJysnJzc5WRkaGMjAzl5uYqJSXFnF9VVaXhw4frzJkz2rZtm9LT0/XWW29pxowZVjcJAADAEssP0rz77rt199131zrP4XAoMzPTo23p0qW6/fbblZ+fr3bt2sntduvVV1/V66+/roEDB0qS1qxZo+joaG3atElDhgzRoUOHlJGRoR07dphPcn355ZeVkJCgTz75RB06dNDGjRt18OBBHTt2TC6XS5L03HPPady4cXr66ae9dlEZAAC4+jX4NVBut1s2m02tW7eWdOEx8ZWVlRo8eLBZ43K5FBcXZz56Pjs7Ww6Hw+Mx+L169ZLD4fCoiYuLM8OTJA0ZMkTl5eUeL3T8tvLycpWUlHh8AADedQ3fDI7LaOp/Fw0aoM6dO6fHH39cycnJ5ohQYWGh/P39FRoa6lEbFRWlwsJCs6a2Z0pERkZ61Fx8t85FoaGh8vf3N2sulZaWZl5T5XA4FB0dfcXbCAD4fi4+E+ns2bNe7gmaoot/F5e+wqapaLB34VVWVur+++9XdXW1Xnzxxe+s//Y7giR5/PeV1HzbnDlzNH36dHO6pKSEEAUAXuLj46PWrVubL8QNDAz8r///xrXDMAydPXtWRUVFat26tXx8fLzdpVo1SICqrKzUyJEjlZeXp3/+858e1yM5nU5VVFSouLjYYxSqqKhIvXv3NmtOnDhRY7knT540R52cTqfHe4akC3fuVVZW1hiZushut8tut9c6DwDQ+JxOpySZIQq4qHXr1ubfR1NU7wHqYnj69NNP9cEHHyg8PNxjfnx8vPz8/JSZmamRI0dKkgoKCrR//34tXLhQkpSQkCC3261du3bp9ttvlyTt3LlTbrfbDFkJCQl6+umnVVBQoLZt20qSNm7cKLvd7vE4fQBA02Wz2dS2bVtFRkaqsrLS291BE+Hn59dkR54ushygSktLdeTIEXM6Ly9Pubm5CgsLk8vl0k9/+lPt2bNH7733nqqqqszrkcLCwuTv7y+Hw6Hx48drxowZCg8PV1hYmGbOnKnOnTubd+V17NhRQ4cO1YQJE7RixQpJ0sSJE5WYmKgOHTpIkgYPHqxOnTopJSVFzz77rL755hvNnDlTEyZM4A48AGhmfHx8mvwPJvBtlgPUhx9+qP79+5vTF68pGjt2rObOnav169dLkrp27erxvQ8++MB8YeKSJUvk6+urkSNHqqysTAMGDNDq1as9/vGsXbtW06ZNM+/WS0pK8nj2lI+Pj/72t79p8uTJuuOOOxQQEKDk5GQtWsR77AAAQMPiZcK8TBgAgGaFlwkDAAA0QwQoAAAAiwhQAAAAFhGgAAAALCJAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAAAsIkABAABYRIACAACwiAAFAABgEQEKAADAIgIUAACARQQoAAAAiwhQAAAAFhGgAAAALCJAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLLAeoLVu2aMSIEXK5XLLZbHrnnXc85huGoblz58rlcikgIED9+vXTgQMHPGrKy8s1depURUREKCgoSElJSfriiy88aoqLi5WSkiKHwyGHw6GUlBSdOnXKoyY/P18jRoxQUFCQIiIiNG3aNFVUVFjdJAAAAEssB6gzZ87o1ltv1bJly2qdv3DhQi1evFjLli3T7t275XQ6NWjQIJ0+fdqsSU1N1bp165Senq5t27aptLRUiYmJqqqqMmuSk5OVm5urjIwMZWRkKDc3VykpKeb8qqoqDR8+XGfOnNG2bduUnp6ut956SzNmzLC6SQAAANYYV0CSsW7dOnO6urracDqdxjPPPGO2nTt3znA4HMZLL71kGIZhnDp1yvDz8zPS09PNmi+//NJo0aKFkZGRYRiGYRw8eNCQZOzYscOsyc7ONiQZH3/8sWEYhrFhwwajRYsWxpdffmnW/OlPfzLsdrvhdrvr1H+3221IMjKXTjG2vzLD/AAAgKbr4u93XX/vG0K9XgOVl5enwsJCDR482Gyz2+3q27evtm/fLknKyclRZWWlR43L5VJcXJxZk52dLYfDoZ49e5o1vXr1ksPh8KiJi4uTy+Uya4YMGaLy8nLl5OTU2r/y8nKVlJR4fAAAAKyq1wBVWFgoSYqKivJoj4qKMucVFhbK399foaGhl62JjIyssfzIyEiPmkvXExoaKn9/f7PmUmlpaeY1VQ6HQ9HR0d9jKwEAwLWuQe7Cs9lsHtOGYdRou9SlNbXVf5+ab5szZ47cbrf5OXbs2GX7BAAAUJt6DVBOp1OSaowAFRUVmaNFTqdTFRUVKi4uvmzNiRMnaiz/5MmTHjWXrqe4uFiVlZU1RqYustvtCgkJ8fgAAABYVa8BKiYmRk6nU5mZmWZbRUWFsrKy1Lt3b0lSfHy8/Pz8PGoKCgq0f/9+syYhIUFut1u7du0ya3bu3Cm32+1Rs3//fhUUFJg1GzdulN1uV3x8fH1uFgAAgAdfq18oLS3VkSNHzOm8vDzl5uYqLCxM7dq1U2pqqubPn6/Y2FjFxsZq/vz5CgwMVHJysiTJ4XBo/PjxmjFjhsLDwxUWFqaZM2eqc+fOGjhwoCSpY8eOGjp0qCZMmKAVK1ZIkiZOnKjExER16NBBkjR48GB16tRJKSkpevbZZ/XNN99o5syZmjBhAiNLAACgQVkOUB9++KH69+9vTk+fPl2SNHbsWK1evVqzZs1SWVmZJk+erOLiYvXs2VMbN25UcHCw+Z0lS5bI19dXI0eOVFlZmQYMGKDVq1fLx8fHrFm7dq2mTZtm3q2XlJTk8ewpHx8f/e1vf9PkyZN1xx13KCAgQMnJyVq0aJH1vQAAAGCBzTAMw9ud8JaSkhI5HA5lLp2ioAC72Z4wnhAGAEBTdfH32+12e+2sE+/CAwAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAAAsIkABAABYRIACAACwiAAFAABgEQEKAADAIgIUAACARQQoAAAAiwhQAAAAFhGgAAAALCJAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAAAsIkABAABYRIACAACwiAAFAABgUb0HqPPnz+tXv/qVYmJiFBAQoBtuuEFPPfWUqqurzRrDMDR37ly5XC4FBASoX79+OnDggMdyysvLNXXqVEVERCgoKEhJSUn64osvPGqKi4uVkpIih8Mhh8OhlJQUnTp1qr43CQAAwEO9B6gFCxbopZde0rJly3To0CEtXLhQzz77rJYuXWrWLFy4UIsXL9ayZcu0e/duOZ1ODRo0SKdPnzZrUlNTtW7dOqWnp2vbtm0qLS1VYmKiqqqqzJrk5GTl5uYqIyNDGRkZys3NVUpKSn1vEgAAgAebYRhGfS4wMTFRUVFRevXVV822n/zkJwoMDNTrr78uwzDkcrmUmpqq2bNnS7ow2hQVFaUFCxZo0qRJcrvdatOmjV5//XWNGjVKknT8+HFFR0drw4YNGjJkiA4dOqROnTppx44d6tmzpyRpx44dSkhI0Mcff6wOHTp8Z19LSkrkcDiUuXSKggLsZnvC+EX1uUsAAEA9uvj77Xa7FRIS4pU+1PsIVJ8+ffT+++/r8OHDkqSPPvpI27Zt07BhwyRJeXl5Kiws1ODBg83v2O129e3bV9u3b5ck5eTkqLKy0qPG5XIpLi7OrMnOzpbD4TDDkyT16tVLDofDrLlUeXm5SkpKPD4AAABW+db3AmfPni23262bb75ZPj4+qqqq0tNPP60HHnhAklRYWChJioqK8vheVFSUjh49atb4+/srNDS0Rs3F7xcWFioyMrLG+iMjI82aS6WlpWnevHlXtoEAAOCaV+8jUH/+85+1Zs0avfHGG9qzZ49ee+01LVq0SK+99ppHnc1m85g2DKNG26Uuramt/nLLmTNnjtxut/k5duxYXTcLAADAVO8jUI899pgef/xx3X///ZKkzp076+jRo0pLS9PYsWPldDolXRhBatu2rfm9oqIic1TK6XSqoqJCxcXFHqNQRUVF6t27t1lz4sSJGus/efJkjdGti+x2u+x2e63zAAAA6qreR6DOnj2rFi08F+vj42M+xiAmJkZOp1OZmZnm/IqKCmVlZZnhKD4+Xn5+fh41BQUF2r9/v1mTkJAgt9utXbt2mTU7d+6U2+02awAAABpCvY9AjRgxQk8//bTatWunW265RXv37tXixYv10EMPSbpw2i01NVXz589XbGysYmNjNX/+fAUGBio5OVmS5HA4NH78eM2YMUPh4eEKCwvTzJkz1blzZw0cOFCS1LFjRw0dOlQTJkzQihUrJEkTJ05UYmJine7AAwAA+L7qPUAtXbpUTz75pCZPnqyioiK5XC5NmjRJv/71r82aWbNmqaysTJMnT1ZxcbF69uypjRs3Kjg42KxZsmSJfH19NXLkSJWVlWnAgAFavXq1fHx8zJq1a9dq2rRp5t16SUlJWrZsWX1vEgAAgId6fw5Uc8JzoAAAaH6uyudAAQAAXO0IUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAAAsIkABAABYRIACAACwiAAFAABgEQEKAADAIgIUAACARQQoAAAAiwhQAAAAFhGgAAAALCJAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAAAsIkABAABYRIACAACwiAAFAABgEQEKAADAIgIUAACARQQoAAAAiwhQAAAAFjVIgPryyy/14IMPKjw8XIGBgeratatycnLM+YZhaO7cuXK5XAoICFC/fv104MABj2WUl5dr6tSpioiIUFBQkJKSkvTFF1941BQXFyslJUUOh0MOh0MpKSk6depUQ2wSAACAqd4DVHFxse644w75+fnp73//uw4ePKjnnntOrVu3NmsWLlyoxYsXa9myZdq9e7ecTqcGDRqk06dPmzWpqalat26d0tPTtW3bNpWWlioxMVFVVVVmTXJysnJzc5WRkaGMjAzl5uYqJSWlvjcJAADAg80wDKM+F/j444/rX//6l7Zu3VrrfMMw5HK5lJqaqtmzZ0u6MNoUFRWlBQsWaNKkSXK73WrTpo1ef/11jRo1SpJ0/PhxRUdHa8OGDRoyZIgOHTqkTp06aceOHerZs6ckaceOHUpISNDHH3+sDh06fGdfS0pK5HA4lLl0ioIC7GZ7wvhFV7obAABAA7n4++12uxUSEuKVPtT7CNT69evVo0cP3XfffYqMjFS3bt308ssvm/Pz8vJUWFiowYMHm212u119+/bV9u3bJUk5OTmqrKz0qHG5XIqLizNrsrOz5XA4zPAkSb169ZLD4TBrLlVeXq6SkhKPDwAAgFX1HqD+85//aPny5YqNjdU//vEPPfzww5o2bZr++Mc/SpIKCwslSVFRUR7fi4qKMucVFhbK399foaGhl62JjIyssf7IyEiz5lJpaWnm9VIOh0PR0dFXtrEAAOCaVO8Bqrq6Wt27d9f8+fPVrVs3TZo0SRMmTNDy5cs96mw2m8e0YRg12i51aU1t9Zdbzpw5c+R2u83PsWPH6rpZAAAApnoPUG3btlWnTp082jp27Kj8/HxJktPplKQao0RFRUXmqJTT6VRFRYWKi4svW3PixIka6z958mSN0a2L7Ha7QkJCPD4AAABW1XuAuuOOO/TJJ594tB0+fFjt27eXJMXExMjpdCozM9OcX1FRoaysLPXu3VuSFB8fLz8/P4+agoIC7d+/36xJSEiQ2+3Wrl27zJqdO3fK7XabNQAAAA3Bt74X+Mtf/lK9e/fW/PnzNXLkSO3atUsrV67UypUrJV047Zaamqr58+crNjZWsbGxmj9/vgIDA5WcnCxJcjgcGj9+vGbMmKHw8HCFhYVp5syZ6ty5swYOHCjpwqjW0KFDNWHCBK1YsUKSNHHiRCUmJtbpDjwAAIDvq94D1G233aZ169Zpzpw5euqppxQTE6Pnn39eo0ePNmtmzZqlsrIyTZ48WcXFxerZs6c2btyo4OBgs2bJkiXy9fXVyJEjVVZWpgEDBmj16tXy8fExa9auXatp06aZd+slJSVp2bJl9b1JAAAAHur9OVDNCc+BAgCg+bkqnwMFAABwtSNAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAAAsIkABAABYRIACAACwiAAFAABgEQEKAADAIgIUAACARQQoAAAAiwhQAAAAFhGgAAAALCJAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAAAsIkABAABY1OABKi0tTTabTampqWabYRiaO3euXC6XAgIC1K9fPx04cMDje+Xl5Zo6daoiIiIUFBSkpKQkffHFFx41xcXFSklJkcPhkMPhUEpKik6dOtXQmwQAAK5xDRqgdu/erZUrV6pLly4e7QsXLtTixYu1bNky7d69W06nU4MGDdLp06fNmtTUVK1bt07p6enatm2bSktLlZiYqKqqKrMmOTlZubm5ysjIUEZGhnJzc5WSktKQmwQAANBwAaq0tFSjR4/Wyy+/rNDQULPdMAw9//zzeuKJJ/TjH/9YcXFxeu2113T27Fm98cYbkiS3261XX31Vzz33nAYOHKhu3bppzZo12rdvnzZt2iRJOnTokDIyMvTKK68oISFBCQkJevnll/Xee+/pk08+aajNAgAAaLgA9cgjj2j48OEaOHCgR3teXp4KCws1ePBgs81ut6tv377avn27JCknJ0eVlZUeNS6XS3FxcWZNdna2HA6Hevbsadb06tVLDofDrLlUeXm5SkpKPD4AAABW+TbEQtPT07Vnzx7t3r27xrzCwkJJUlRUlEd7VFSUjh49atb4+/t7jFxdrLn4/cLCQkVGRtZYfmRkpFlzqbS0NM2bN8/6BgEAAHxLvY9AHTt2TI8++qjWrFmjli1b/tc6m83mMW0YRo22S11aU1v95ZYzZ84cud1u83Ps2LHLrg8AAKA29R6gcnJyVFRUpPj4ePn6+srX11dZWVl64YUX5Ovra448XTpKVFRUZM5zOp2qqKhQcXHxZWtOnDhRY/0nT56sMbp1kd1uV0hIiMcHAADAqnoPUAMGDNC+ffuUm5trfnr06KHRo0crNzdXN9xwg5xOpzIzM83vVFRUKCsrS71795YkxcfHy8/Pz6OmoKBA+/fvN2sSEhLkdru1a9cus2bnzp1yu91mDQAAQEOo92uggoODFRcX59EWFBSk8PBwsz01NVXz589XbGysYmNjNX/+fAUGBio5OVmS5HA4NH78eM2YMUPh4eEKCwvTzJkz1blzZ/Oi9I4dO2ro0KGaMGGCVqxYIUmaOHGiEhMT1aFDh/reLAAAAFODXET+XWbNmqWysjJNnjxZxcXF6tmzpzZu3Kjg4GCzZsmSJfL19dXIkSNVVlamAQMGaPXq1fLx8TFr1q5dq2nTppl36yUlJWnZsmWNvj0AAODaYjMMw/B2J7ylpKREDodDmUunKCjAbrYnjF/kxV4BAIDLufj77Xa7vXY9M+/CAwAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAAAsIkABAABYRIACAACwiAAFAABgEQEKAADAIgIUAACARQQoAAAAiwhQAAAAFhGgAAAALCJAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACwiQAEAAFjk6+0ONEkfpNVs6z+n8fsBAACaJEagAAAALCJAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACyq9yeRp6Wl6e2339bHH3+sgIAA9e7dWwsWLFCHDh3MGsMwNG/ePK1cuVLFxcXq2bOnfv/73+uWW24xa8rLyzVz5kz96U9/UllZmQYMGKAXX3xR1113nVlTXFysadOmaf369ZKkpKQkLV26VK1bt76ibcj+z9c12nacP1yj7Ze+b9X8Mk8sBwDgqlfvI1BZWVl65JFHtGPHDmVmZur8+fMaPHiwzpw5Y9YsXLhQixcv1rJly7R79245nU4NGjRIp0+fNmtSU1O1bt06paena9u2bSotLVViYqKqqqrMmuTkZOXm5iojI0MZGRnKzc1VSkpKfW+SJKlX/soaHwAAcG2yGYZhNOQKTp48qcjISGVlZenOO++UYRhyuVxKTU3V7NmzJV0YbYqKitKCBQs0adIkud1utWnTRq+//rpGjRolSTp+/Liio6O1YcMGDRkyRIcOHVKnTp20Y8cO9ezZU5K0Y8cOJSQk6OOPP/YY8fpvSkpK5HA4lLl0ioIC7Ja3LeGG8JqN9TwCtSSzlpGvQTfV6zoAAGhOLv5+u91uhYSEeKUPDf4yYbfbLUkKCwuTJOXl5amwsFCDBw82a+x2u/r27avt27dr0qRJysnJUWVlpUeNy+VSXFyctm/friFDhig7O1sOh8MMT5LUq1cvORwObd++vdYAVV5ervLycnO6pKSk3re3VvX8cmJCFQAA3tWgAcowDE2fPl19+vRRXFycJKmwsFCSFBUV5VEbFRWlo0ePmjX+/v4KDQ2tUXPx+4WFhYqMjKyxzsjISLPmUmlpaZo3b96VbdR3qDXcXMFeru1U4Y52E+u23rqGqnoOeAAAXO0aNEBNmTJF//73v7Vt27Ya82w2m8e0YRg12i51aU1t9Zdbzpw5czR9+nRzuqSkRNHR0Zdd5+XUdrG52n3vxXlNbduR0N8LHamrKwh8jN4BAOpDgwWoqVOnav369dqyZYvHnXNOp1PShRGktm3bmu1FRUXmqJTT6VRFRYWKi4s9RqGKiorUu3dvs+bEiRM11nvy5Mkao1sX2e122e3Wr3VqELWEgCXnf1KjrdcVrKKhw0JTCiNNqS8AgKtfvQcowzA0depUrVu3Tps3b1ZMTIzH/JiYGDmdTmVmZqpbt26SpIqKCmVlZWnBggWSpPj4ePn5+SkzM1MjR46UJBUUFGj//v1auHChJCkhIUFut1u7du3S7bffLknauXOn3G63GbKastpGfXrpKrizr46jQ9mvzqzRljB+UZ1WUdeRv9pCFQAA9aHeA9QjjzyiN954Q++++66Cg4PN65EcDocCAgJks9mUmpqq+fPnKzY2VrGxsZo/f74CAwOVnJxs1o4fP14zZsxQeHi4wsLCNHPmTHXu3FkDBw6UJHXs2FFDhw7VhAkTtGLFCknSxIkTlZiYWKc78BpTrT/4V4FaH+VQ252JV6K2QFbHvtR2rVhtar9ujWd8AQD+u3oPUMuXL5ck9evXz6N91apVGjdunCRp1qxZKisr0+TJk80HaW7cuFHBwcFm/ZIlS+Tr66uRI0eaD9JcvXq1fHx8zJq1a9dq2rRp5t16SUlJWrZsWX1vkiWN8XyouoaF2vtSt1GeGuoYZK5EraNS9R3IrmKcxgSAxtPgz4Fqyq70OVBNSV0D1Petq21EptaLz+sYeLw1KlfXUam6jkBdSWip78BT11OWhCoAzd018RwoNI6m8mT0q/V05RWrdQSv5k0DTcWVXD9GQANwLSBAXWOaStBqSup8LVddQ9AVnO6s71GpWrftg4Z9gn5TGpVrSq7mbQOuRQQoXFPqGiDrOpJW652TtYSvK7nrcklmzdOOtZ1i7JX//Uf/GvyORR7WCuAqQ4BCnXBqru6a+r7yxgNgr+Rhrd66tqupjxjV50gfby0ArCNAoVac6ms6GuNxEZeuo64X2zeGK3lERb3zVoBoItfQNbu3FgANiAAFoIZGCS21hIIrOQ1Z13XUd+Dx1khVvT6mpKmr63WFjIahERGggGaoKZ8m9NboZV1f5N0YT8G/khGj+n4heVPW1E+T1tXVsh2w5ir9ZwmgvtX3qNSVhMC69qXO67iCwFPXU6yNMWLUVH7IvfYapbqOODb1a7nq+K7U2hDcGg8BCsBVoTlet1fbaJjq+VRpQ4eZel9+I7z1oDZXEj6vqdOpMBGgAKAJuZKbBuoaIr1xEf6VjBrW9oaDuo4uJsg7gayu6vuUbVMZhbwiTX2E8P8QoACgFlcyotWUr1GrTW0/us1xRK82dQ1kdX3obG2n0nrVst667tPavlvXU8BN6W7ZGppJCLoSBCgA39vV/CN7NavPx1ZcyQ+7t/5+mvrxrvf+XRpm6hpkvBSCmsvjMghQANDENfUf/EtdzcG6rm8QaAzfN7zWeu1dHdUWZGq9s7WWUbSr4vTitxCgAOAa19QDT3MLkFLT2qf1uf/qGr7qHj6b78X2BCgAAK4STSm41cWVjIZ5WwtvdwAAAKC5IUABAABYRIACAACwiAAFAABgEQEKAADAIgIUAACARQQoAAAAiwhQAAAAFhGgAAAALCJAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAAAsIkABAABYRIACAACwqNkHqBdffFExMTFq2bKl4uPjtXXrVm93CQAAXOWadYD685//rNTUVD3xxBPau3evfvjDH+ruu+9Wfn6+t7sGAACuYs06QC1evFjjx4/Xz3/+c3Xs2FHPP/+8oqOjtXz5cm93DQAAXMV8vd2B76uiokI5OTl6/PHHPdoHDx6s7du31/qd8vJylZeXm9Nut1uSdKasouE6CgAArkhJSUmt04ZheKM7kppxgPrqq69UVVWlqKgoj/aoqCgVFhbW+p20tDTNmzevRvu9s1Y2SB8BAEA9mLqs1uavv/5aDoejkTtzQbMNUBfZbDaPacMwarRdNGfOHE2fPt2cPnXqlNq3b6/8/HyvHQBcUFJSoujoaB07dkwhISHe7s41jWPRtHA8mg6ORdPhdrvVrl07hYWFea0PzTZARUREyMfHp8ZoU1FRUY1RqYvsdrvsdnuNdofDwT+GJiIkJIRj0URwLJoWjkfTwbFoOlq08N6l3M32InJ/f3/Fx8crMzPToz0zM1O9e/f2Uq8AAMC1oNmOQEnS9OnTlZKSoh49eighIUErV65Ufn6+Hn74YW93DQAAXMWadYAaNWqUvv76az311FMqKChQXFycNmzYoPbt29fp+3a7Xb/5zW9qPa2HxsWxaDo4Fk0Lx6Pp4Fg0HU3hWNgMb94DCAAA0Aw122ugAAAAvIUABQAAYBEBCgAAwCICFAAAgEXXdIB68cUXFRMTo5YtWyo+Pl5bt271dpeatbS0NN12220KDg5WZGSk7r33Xn3yySceNYZhaO7cuXK5XAoICFC/fv104MABj5ry8nJNnTpVERERCgoKUlJSkr744guPmuLiYqWkpMjhcMjhcCglJUWnTp1q6E1sltLS0mSz2ZSammq2cRwa15dffqkHH3xQ4eHhCgwMVNeuXZWTk2PO53g0jvPnz+tXv/qVYmJiFBAQoBtuuEFPPfWUqqurzRqORcPYsmWLRowYIZfLJZvNpnfeecdjfmPu9/z8fI0YMUJBQUGKiIjQtGnTVFHxPd6Ja1yj0tPTDT8/P+Pll182Dh48aDz66KNGUFCQcfToUW93rdkaMmSIsWrVKmP//v1Gbm6uMXz4cKNdu3ZGaWmpWfPMM88YwcHBxltvvWXs27fPGDVqlNG2bVujpKTErHn44YeNH/zgB0ZmZqaxZ88eo3///satt95qnD9/3qwZOnSoERcXZ2zfvt3Yvn27ERcXZyQmJjbq9jYHu3btMq6//nqjS5cuxqOPPmq2cxwazzfffGO0b9/eGDdunLFz504jLy/P2LRpk3HkyBGzhuPROH73u98Z4eHhxnvvvWfk5eUZb775ptGqVSvj+eefN2s4Fg1jw4YNxhNPPGG89dZbhiRj3bp1HvMba7+fP3/eiIuLM/r372/s2bPHyMzMNFwulzFlyhTL23TNBqjbb7/dePjhhz3abr75ZuPxxx/3Uo+uPkVFRYYkIysryzAMw6iurjacTqfxzDPPmDXnzp0zHA6H8dJLLxmGYRinTp0y/Pz8jPT0dLPmyy+/NFq0aGFkZGQYhmEYBw8eNCQZO3bsMGuys7MNScbHH3/cGJvWLJw+fdqIjY01MjMzjb59+5oBiuPQuGbPnm306dPnv87neDSe4cOHGw899JBH249//GPjwQcfNAyDY9FYLg1QjbnfN2zYYLRo0cL48ssvzZo//elPht1uN9xut6XtuCZP4VVUVCgnJ0eDBw/2aB88eLC2b9/upV5dfdxutySZL3vMy8tTYWGhx3632+3q27evud9zcnJUWVnpUeNyuRQXF2fWZGdny+FwqGfPnmZNr1695HA4OH7f8sgjj2j48OEaOHCgRzvHoXGtX79ePXr00H333afIyEh169ZNL7/8sjmf49F4+vTpo/fff1+HDx+WJH300Ufatm2bhg0bJolj4S2Nud+zs7MVFxcnl8tl1gwZMkTl5eUep9Xrolk/ifz7+uqrr1RVVVXjpcNRUVE1Xk6M78cwDE2fPl19+vRRXFycJJn7trb9fvToUbPG399foaGhNWoufr+wsFCRkZE11hkZGcnx+z/p6enas2ePdu/eXWMex6Fx/ec//9Hy5cs1ffp0/b//9/+0a9cuTZs2TXa7XWPGjOF4NKLZs2fL7Xbr5ptvlo+Pj6qqqvT000/rgQcekMS/DW9pzP1eWFhYYz2hoaHy9/e3fGyuyQB1kc1m85g2DKNGG76fKVOm6N///re2bdtWY9732e+X1tRWz/G74NixY3r00Ue1ceNGtWzZ8r/WcRwaR3V1tXr06KH58+dLkrp166YDBw5o+fLlGjNmjFnH8Wh4f/7zn7VmzRq98cYbuuWWW5Sbm6vU1FS5XC6NHTvWrONYeEdj7ff6OjbX5Cm8iIgI+fj41EibRUVFNZIprJs6darWr1+vDz74QNddd53Z7nQ6Jemy+93pdKqiokLFxcWXrTlx4kSN9Z48eZLjpwtD3UVFRYqPj5evr698fX2VlZWlF154Qb6+vuY+4jg0jrZt26pTp04ebR07dlR+fr4k/l00pscee0yPP/647r//fnXu3FkpKSn65S9/qbS0NEkcC29pzP3udDprrKe4uFiVlZWWj801GaD8/f0VHx+vzMxMj/bMzEz17t3bS71q/gzD0JQpU/T222/rn//8p2JiYjzmx8TEyOl0euz3iooKZWVlmfs9Pj5efn5+HjUFBQXav3+/WZOQkCC3261du3aZNTt37pTb7eb4SRowYID27dun3Nxc89OjRw+NHj1aubm5uuGGGzgOjeiOO+6o8TiPw4cPmy89599F4zl79qxatPD82fPx8TEfY8Cx8I7G3O8JCQnav3+/CgoKzJqNGzfKbrcrPj7eWsctXXJ+Fbn4GINXX33VOHjwoJGammoEBQUZn3/+ube71mz94he/MBwOh7F582ajoKDA/Jw9e9aseeaZZwyHw2G8/fbbxr59+4wHHnig1ltVr7vuOmPTpk3Gnj17jLvuuqvWW1W7dOliZGdnG9nZ2Ubnzp2v6VuEv8u378IzDI5DY9q1a5fh6+trPP3008ann35qrF271ggMDDTWrFlj1nA8GsfYsWONH/zgB+ZjDN5++20jIiLCmDVrllnDsWgYp0+fNvbu3Wvs3bvXkGQsXrzY2Lt3r/nooMba7xcfYzBgwABjz549xqZNm4zrrruOxxhY9fvf/95o37694e/vb3Tv3t283R7fj6RaP6tWrTJrqqurjd/85jeG0+k07Ha7ceeddxr79u3zWE5ZWZkxZcoUIywszAgICDASExON/Px8j5qvv/7aGD16tBEcHGwEBwcbo0ePNoqLixthK5unSwMUx6Fx/e///q8RFxdn2O124+abbzZWrlzpMZ/j0ThKSkqMRx991GjXrp3RsmVL44YbbjCeeOIJo7y83KzhWDSMDz74oNbfh7FjxxqG0bj7/ejRo8bw4cONgIAAIywszJgyZYpx7tw5y9tkMwzDsDZmBQAAcG27Jq+BAgAAuBIEKAAAAIsIUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAAAs+v8BJ9Lvn1t2xQAAAABJRU5ErkJggg==\n",
+ "text/plain": [
+ "<Figure size 640x480 with 1 Axes>"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ }
+ ],
+ "source": [
+ "plt.hist( peaks_closest_upstream_nodir['distance'], np.arange(0, 1e4, 100), alpha=0.5, label=\"upstream\");\n",
+ "plt.hist( peaks_closest_downstream_nodir['distance'], np.arange(0, 1e4, 100), alpha=0.5, label=\"downstream\");\n",
+ "plt.xlim( [0, 1e4] )\n",
+ "plt.legend()"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "4f130a50",
+ "metadata": {},
+ "source": [
+ "## Ignore upstream/downstream peaks from genes (strand-aware version)\n",
+ "\n",
+ "More biologically relevant approach will be to **define upstream/downstream by strand of the gene**.\n",
+ "CTCF upstream of transcription start site might play different role than CTCF after transcription end site. \n",
+ "\n",
+ "`bioframe.closest` has the parameter `direction_col` to control for that: \n",
+ "\n",
+ "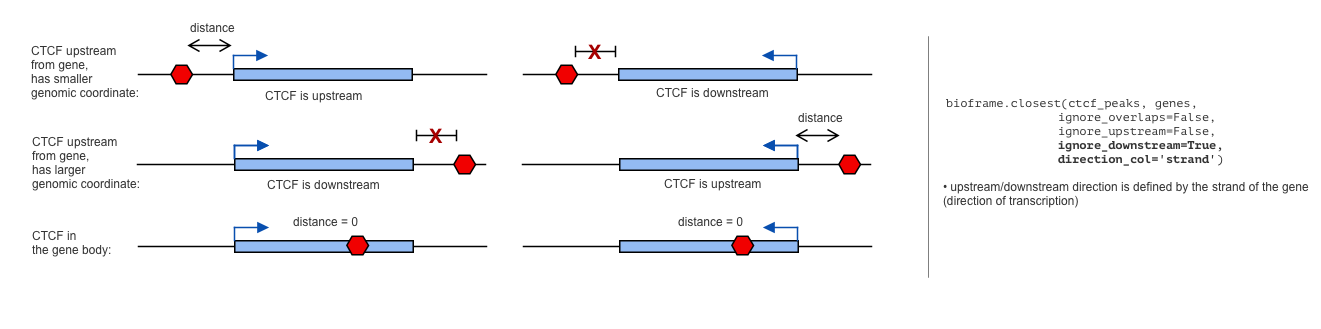\n"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 90,
+ "id": "b47d83fa",
+ "metadata": {},
+ "outputs": [],
+ "source": [
+ "# Note that \"strand\" here is the column name in genes table:\n",
+ "peaks_closest_upstream_dir = bioframe.closest(genes, ctcf_peaks, \n",
+ " ignore_overlaps=False,\n",
+ " ignore_upstream=False,\n",
+ " ignore_downstream=True,\n",
+ " direction_col='strand')\n",
+ "\n",
+ "peaks_closest_downstream_dir = bioframe.closest(genes, ctcf_peaks, \n",
+ " ignore_overlaps=False,\n",
+ " ignore_upstream=True,\n",
+ " ignore_downstream=False,\n",
+ " direction_col='strand')"
+ ]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 96,
+ "id": "f18e706a",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "text/plain": [
+ "<matplotlib.legend.Legend at 0x7fd14a781610>"
+ ]
+ },
+ "execution_count": 96,
+ "metadata": {},
+ "output_type": "execute_result"
+ },
+ {
+ "data": {
+ "image/png": "iVBORw0KGgoAAAANSUhEUgAAAlAAAAGdCAYAAADdfE2yAAAAOXRFWHRTb2Z0d2FyZQBNYXRwbG90bGliIHZlcnNpb24zLjQuMywgaHR0cHM6Ly9tYXRwbG90bGliLm9yZy/MnkTPAAAACXBIWXMAAA9hAAAPYQGoP6dpAAA+HElEQVR4nO3de1hVZf7//xdy2ALiloOw3aVGnxjTwFRsFHWSUtEUyWkmLQx1ctTGlBg1zW9TY80kHUz9pJ9Mq9FGLWaasqyMEZtEHUENpUItqyFRA7HCjQcExPX7w5/rmi1krNwc1OfjuvZ1te/1Xmvf91p47Vf3OmwvwzAMAQAAoN5aNHUHAAAALjUEKAAAAIsIUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAin6buQFM6c+aMvvnmGwUFBcnLy6upuwMAAOrBMAwdO3ZMTqdTLVo0zVzQFR2gvvnmG7Vv376puwEAAH6CAwcO6Oqrr26Sz76iA1RQUJCkswegdevWTdwbAABQH+Xl5Wrfvr35Pd4UrugAde60XevWrQlQAABcYpry8hsuIgcAALCIAAUAAGARAQoAAMCiK/oaKABA0zMMQ6dPn1ZNTU1TdwXNhLe3t3x8fJr1I4YsB6hNmzbpmWeeUV5enoqLi7VmzRqNGDGiztpJkyZp2bJlWrBggdLS0sz2yspKzZgxQ6+99poqKio0YMAAPf/88263IpaVlSk1NVVr166VJCUlJWnRokVq06aNWVNUVKT7779f//rXv+Tv76/k5GTNmzdPfn5+VocFAGgCVVVVKi4u1smTJ5u6K2hmAgIC1K5du2b7nW45QJ04cUI33nijfvOb3+hXv/rVD9a99dZb2rZtm5xOZ61laWlpeuedd5SRkaHQ0FBNnz5diYmJysvLk7e3tyQpOTlZBw8eVGZmpiRp4sSJSklJ0TvvvCNJqqmp0bBhw9S2bVtt2bJF3333ncaOHSvDMLRo0SKrwwIANLIzZ86osLBQ3t7ecjqd8vPza9YzDmgchmGoqqpKR44cUWFhoaKioprsYZkXZFwEScaaNWtqtR88eNC46qqrjIKCAqNjx47GggULzGVHjx41fH19jYyMDLPt0KFDRosWLYzMzEzDMAxjz549hiQjNzfXrMnJyTEkGZ999plhGIaxbt06o0WLFsahQ4fMmtdee82w2WyGy+WqV/9dLpchqd71AADPqaioMPbs2WOcOHGiqbuCZujEiRPGnj17jIqKilrLmsP3t8cj3ZkzZ5SSkqIHH3xQN9xwQ63leXl5qq6uVkJCgtnmdDoVHR2trVu3SpJycnJkt9vVq1cvs6Z3796y2+1uNdHR0W4zXIMHD1ZlZaXy8vLq7FtlZaXKy8vdXgCAptUsZxfQ5Jr734XHe/fUU0/Jx8dHqampdS4vKSmRn5+fgoOD3dojIiJUUlJi1oSHh9daNzw83K0mIiLCbXlwcLD8/PzMmvOlp6fLbrebL37GBQAA/BQeDVB5eXn63//9X61YscLyeWzDMNzWqWv9n1Lz32bPni2Xy2W+Dhw4YKmPAAAAkocfY7B582aVlpaqQ4cOZltNTY2mT5+uhQsX6uuvv5bD4VBVVZXKysrcZqFKS0vVp08fSZLD4dDhw4drbf/IkSPmrJPD4dC2bdvclpeVlam6urrWzNQ5NptNNpvtoscJAGg4C7L2Nern/X7Qzxr1836Ml5fXBe9wR/Pg0RmolJQUffLJJ8rPzzdfTqdTDz74oP75z39KkmJjY+Xr66usrCxzveLiYhUUFJgBKi4uTi6XS9u3bzdrtm3bJpfL5VZTUFCg4uJis2b9+vWy2WyKjY315LAAAGhWqqurm7oLVzzLAer48eNmOJKkwsJC5efnq6ioSKGhoYqOjnZ7+fr6yuFwqFOnTpIku92u8ePHa/r06frggw+0a9cu3XPPPYqJidHAgQMlSZ07d9aQIUM0YcIE5ebmKjc3VxMmTFBiYqK5nYSEBHXp0kUpKSnatWuXPvjgA82YMUMTJkzgh4EBAA3mmmuu0cKFC93aunXrpjlz5kg6O4O0ZMkS3XbbbfL391dkZKRef/11s7aqqkpTpkxRu3bt1LJlS11zzTVKT083ty1Jv/zlL+Xl5WW+nzNnjrp166a//OUvuvbaa2Wz2WQYhlwulyZOnKjw8HC1bt1at956qz7++GPzs7766ivdfvvtioiIUKtWrXTTTTdpw4YNtcbz5z//WWPGjFGrVq3UsWNHvf322zpy5Ihuv/12tWrVSjExMfroo488uyMvcZYD1EcffaTu3bure/fukqRp06ape/fuevTRR+u9jQULFmjEiBEaOXKk+vbtq4CAAL3zzjvmM6AkafXq1YqJiVFCQoISEhLUtWtXrVy50lzu7e2t9957Ty1btlTfvn01cuRIjRgxQvPmzbM6JAAAPOqRRx7Rr371K3388ce65557dPfdd2vv3r2SpOeee05r167V3//+d33++edatWqVGZR27NghSVq+fLmKi4vN95L05Zdf6u9//7veeOMNcxJj2LBhKikp0bp165SXl6cePXpowIAB+v777yWdnfQYOnSoNmzYoF27dmnw4MEaPny4ioqK3Pq7YMEC9e3bV7t27dKwYcOUkpKiMWPG6J577tHOnTt13XXXacyYMTIMo4H33KXD8jVQ8fHxlnbg119/XautZcuWWrRo0QUfeBkSEqJVq1ZdcNsdOnTQu+++W+++/JD/+9eXahnYynzf3M6HAwAuLXfeead++9vfSpL+9Kc/KSsrS4sWLdLzzz+voqIiRUVFqV+/fvLy8lLHjh3N9dq2bStJatOmjRwOh9s2q6qqtHLlSrPmX//6lz799FOVlpaa1/fOmzdPb731lv7xj39o4sSJuvHGG3XjjTea2/jzn/+sNWvWaO3atZoyZYrZPnToUE2aNEmS9Oijj2rJkiW66aabdOedd0qSZs2apbi4OB0+fLhWv65UzfshCwAAXILi4uJqvT83AzVu3Djl5+erU6dOSk1N1fr16+u1zY4dO5rhSTp75/vx48cVGhqqVq1ama/CwkJ99dVXks7+esjMmTPVpUsXtWnTRq1atdJnn31Wawaqa9eu5n+fuxErJiamVltpaWl9d8Fljx8TBgDAghYtWtQ6E1Ofi7rPPWKnR48eKiws1Pvvv68NGzZo5MiRGjhwoP7xj39ccP3AwEC392fOnFG7du20cePGWrXnfjf23E1c8+bN03XXXSd/f3/9+te/VlVVlVu9r69vrX7W1XbmzJkfHeeVggAFAIAFbdu2dbsDvLy8XIWFhW41ubm5GjNmjNv7c9cOS1Lr1q01atQojRo1Sr/+9a81ZMgQff/99woJCZGvr69qamp+tB89evRQSUmJfHx8zGuozrd582aNGzdOv/zlLyWdvSaqrktrYB0BCgAAC2699VatWLFCw4cPV3BwsB555BG3m6Ak6fXXX1fPnj3Vr18/rV69Wtu3b9fLL78s6ewF2+3atVO3bt3UokULvf7663I4HOas0TXXXKMPPvhAffv2lc1mq/XLHecMHDhQcXFxGjFihJ566il16tRJ33zzjdatW6cRI0aoZ8+euu666/Tmm29q+PDh8vLy0iOPPMIskodwDRQAABbMnj1bN998sxITEzV06FCNGDFC//M//+NW89hjjykjI0Ndu3bVK6+8otWrV6tLly6SpFatWumpp55Sz549ddNNN+nrr7/WunXrzN9+e/bZZ5WVlaX27du7zVqdz8vLS+vWrdPNN9+se++9Vz/72c9011136euvvzavWVqwYIGCg4PVp08fDR8+XIMHD1aPHj0aaM9cWbyMK/iexPLyctntds1dk8ddeADQyE6dOqXCwkJFRkaqZcuWTd0dj+FJ4p5xob+Pc9/fLperyZ79yAwUAACARQQoAAAAi7iIHAAAD7qCr4y5ojADBQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACwiQAEA4AHx8fFKS0tr6m6gkfAcKABA8/JheuN+3i2zG/fzGtnXX3+tyMhI7dq1S926dWvq7lw2mIECAACqqqpq6i5cUghQAABYdOLECY0ZM0atWrVSu3bt9Oyzz7otLysr05gxYxQcHKyAgADddttt+uKLLySdfVJ527Zt9cYbb5j13bp1U3h4uPk+JydHvr6+On78uKSzP1D80ksv6Ze//KUCAgIUFRWltWvXun3e6NGj1bZtW/n7+ysqKkrLly+XJEVGRkqSunfvLi8vL8XHx0uSxo0bpxEjRig9PV1Op1M/+9nPJEmHDh3SqFGjFBwcrNDQUN1+++36+uuvzc/asWOHBg0apLCwMNntdvXv3187d+50G7+Xl5eWLl2qxMREBQQEqHPnzsrJydGXX36p+Ph4BQYGKi4uTl999dXFHIYmRYACAMCiBx98UB9++KHWrFmj9evXa+PGjcrLyzOXjxs3Th999JHWrl2rnJwcGYahoUOHqrq6Wl5eXrr55pu1ceNGSWfDz549e1RdXa09e/ZIkjZu3KjY2Fi1atXK3OZjjz2mkSNH6pNPPtHQoUM1evRoff/995KkRx55RHv27NH777+vvXv3asmSJQoLC5Mkbd++XZK0YcMGFRcX68033zS3+cEHH2jv3r3KysrSu+++q5MnT+qWW25Rq1attGnTJm3ZskWtWrXSkCFDzBmqY8eOaezYsdq8ebNyc3MVFRWloUOH6tixY2776E9/+pPGjBmj/Px8XX/99UpOTtakSZM0e/ZsffTRR5KkKVOmePKwNCqugQIAwILjx4/r5Zdf1l//+lcNGjRIkvTKK6/o6quvliR98cUXWrt2rf7973+rT58+kqTVq1erffv2euutt3TnnXcqPj5ey5YtkyRt2rRJN954ozp06KCNGzeqS5cu2rhxozlTdM64ceN09913S5Lmzp2rRYsWafv27RoyZIiKiorUvXt39ezZU5J0zTXXmOu1bdtWkhQaGiqHw+G2zcDAQL300kvy8/OTJP3lL39RixYt9NJLL8nLy0uStHz5crVp00YbN25UQkKCbr31VrdtLF26VMHBwcrOzlZiYqLZ/pvf/EYjR46UJM2aNUtxcXF65JFHNHjwYEnSAw88oN/85jdWd3+zwQwUAAAWfPXVV6qqqlJcXJzZFhISok6dOkmS9u7dKx8fH/Xq1ctcHhoaqk6dOmnv3r2Szt6xt3v3bn377bfKzs5WfHy84uPjlZ2drdOnT2vr1q3q37+/2+d27drV/O/AwEAFBQWptLRUkvS73/1OGRkZ6tatm2bOnKmtW7fWaywxMTFmeJKkvLw8ffnllwoKClKrVq3UqlUrhYSE6NSpU+bpttLSUt1333362c9+JrvdLrvdruPHj6uoqOgH+xsREWF+3n+3nTp1SuXl5fXqa3PDDBQAABYYhvGTlhuGYc7qREdHKzQ0VNnZ2crOztbjjz+u9u3b64knntCOHTtUUVGhfv36ua3v6+vr9t7Ly0tnzpyRJN12223av3+/3nvvPW3YsEEDBgzQ/fffr3nz5l2wr4GBgW7vz5w5o9jYWK1evbpW7bmZrHHjxunIkSNauHChOnbsKJvNpri4uFoXof93f8+Nu662c2O41DADBQCABdddd518fX2Vm5trtpWVlWnfvn2SpC5duuj06dPatm2bufy7777Tvn371LlzZ0kyr4N6++23VVBQoF/84heKiYlRdXW1XnjhBfXo0UNBQUGW+tW2bVuNGzdOq1at0sKFC81ThOdmmGpqan50Gz169NAXX3yh8PBwXXfddW4vu90uSdq8ebNSU1M1dOhQ3XDDDbLZbPr2228t9fVyQIACAMCCVq1aafz48XrwwQf1wQcfqKCgQOPGjVOLFme/UqOionT77bdrwoQJ2rJliz7++GPdc889uuqqq3T77beb24mPj9err76qrl27qnXr1maoWr16da3rn37Mo48+qrfffltffvmldu/erXfffdcMa+Hh4fL391dmZqYOHz4sl8v1g9sZPXq0wsLCdPvtt2vz5s0qLCxUdna2HnjgAR08eFDS2QC5cuVK7d27V9u2bdPo0aPl7+9vcS9e+ghQkm46uFy9i5aZLwAALuSZZ57RzTffrKSkJA0cOFD9+vVTbGysuXz58uWKjY1VYmKi4uLiZBiG1q1b53YK65ZbblFNTY1bWOrfv79qampqXf/0Y/z8/DR79mx17dpVN998s7y9vZWRkSFJ8vHx0XPPPaelS5fK6XS6hbjzBQQEaNOmTerQoYPuuOMOde7cWffee68qKirUunVrSWcvNC8rK1P37t2VkpKi1NRUt0cwXCm8jB87mXsZKy8vl91uV9aiKQr0t5ntceMvfM4YAHDxTp06pcLCQkVGRqply5ZN3R00Mxf6+zj3/e1yucxg19iYgQIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAgCZ1Bd8Mjgto7n8XBCgAQJM490ykkydPNnFP0Byd+7s4/ydsmgt+Cw8A0CS8vb3Vpk0b8wdxAwICzN9Hw5XLMAydPHlSpaWlatOmjby9vZu6S3UiQAEAmozD4ZAkM0QB57Rp08b8+2iOCFAAgCbj5eWldu3aKTw8XNXV1U3dHTQTvr6+zXbm6RwCFACgyXl7ezf7L0zgv3EROQAAgEUEKAAAAIssB6hNmzZp+PDhcjqd8vLy0ltvvWUuq66u1qxZsxQTE6PAwEA5nU6NGTNG33zzjds2KisrNXXqVIWFhSkwMFBJSUk6ePCgW01ZWZlSUlJkt9tlt9uVkpKio0ePutUUFRVp+PDhCgwMVFhYmFJTU1VVVWV1SAAAAJZYDlAnTpzQjTfeqMWLF9dadvLkSe3cuVOPPPKIdu7cqTfffFP79u1TUlKSW11aWprWrFmjjIwMbdmyRcePH1diYqJqamrMmuTkZOXn5yszM1OZmZnKz89XSkqKubympkbDhg3TiRMntGXLFmVkZOiNN97Q9OnTrQ4JAADAEi/jIh716eXlpTVr1mjEiBE/WLNjxw79/Oc/1/79+9WhQwe5XC61bdtWK1eu1KhRoyRJ33zzjdq3b69169Zp8ODB2rt3r7p06aLc3Fz16tVLkpSbm6u4uDh99tln6tSpk95//30lJibqwIEDcjqdkqSMjAyNGzdOpaWlat269Y/2v7y8XHa7XVmLpijQ32a2x42f91N3CQAAaGDnvr9dLle9vu8bQoNfA+VyueTl5aU2bdpIkvLy8lRdXa2EhASzxul0Kjo6Wlu3bpUk5eTkyG63m+FJknr37i273e5WEx0dbYYnSRo8eLAqKyuVl5dXZ18qKytVXl7u9gIAALCqQQPUqVOn9NBDDyk5OdlMiCUlJfLz81NwcLBbbUREhEpKSsya8PDwWtsLDw93q4mIiHBbHhwcLD8/P7PmfOnp6eY1VXa7Xe3bt7/oMQIAgCtPgwWo6upq3XXXXTpz5oyef/75H603DMPtEf51Pc7/p9T8t9mzZ8vlcpmvAwcO1GcoAAAAbhokQFVXV2vkyJEqLCxUVlaW2/lJh8OhqqoqlZWVua1TWlpqzig5HA4dPny41naPHDniVnP+TFNZWZmqq6trzUydY7PZ1Lp1a7cXAACAVR4PUOfC0xdffKENGzYoNDTUbXlsbKx8fX2VlZVlthUXF6ugoEB9+vSRJMXFxcnlcmn79u1mzbZt2+RyudxqCgoKVFxcbNasX79eNptNsbGxnh4WAACAyfJPuRw/flxffvml+b6wsFD5+fkKCQmR0+nUr3/9a+3cuVPvvvuuampqzFmikJAQ+fn5yW63a/z48Zo+fbpCQ0MVEhKiGTNmKCYmRgMHDpQkde7cWUOGDNGECRO0dOlSSdLEiROVmJioTp06SZISEhLUpUsXpaSk6JlnntH333+vGTNmaMKECcwsAQCABmU5QH300Ue65ZZbzPfTpk2TJI0dO1Zz5szR2rVrJUndunVzW+/DDz9UfHy8JGnBggXy8fHRyJEjVVFRoQEDBmjFihVuv4O0evVqpaammnfrJSUluT17ytvbW++9954mT56svn37yt/fX8nJyZo3j0cQAACAhnVRz4G61PEcKAAALj1XxHOgAAAALjcEKAAAAIsIUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAAAsIkABAABYRIACAACwiAAFAABgEQEKAADAIgIUAACARQQoAAAAiwhQAAAAFhGgAAAALCJAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAAAsIkABAABYRIACAACwiAAFAABgEQEKAADAIssBatOmTRo+fLicTqe8vLz01ltvuS03DENz5syR0+mUv7+/4uPjtXv3breayspKTZ06VWFhYQoMDFRSUpIOHjzoVlNWVqaUlBTZ7XbZ7XalpKTo6NGjbjVFRUUaPny4AgMDFRYWptTUVFVVVVkdEgAAgCWWA9SJEyd04403avHixXUuf/rppzV//nwtXrxYO3bskMPh0KBBg3Ts2DGzJi0tTWvWrFFGRoa2bNmi48ePKzExUTU1NWZNcnKy8vPzlZmZqczMTOXn5yslJcVcXlNTo2HDhunEiRPasmWLMjIy9MYbb2j69OlWhwQAAGCNcREkGWvWrDHfnzlzxnA4HMaTTz5ptp06dcqw2+3GCy+8YBiGYRw9etTw9fU1MjIyzJpDhw4ZLVq0MDIzMw3DMIw9e/YYkozc3FyzJicnx5BkfPbZZ4ZhGMa6deuMFi1aGIcOHTJrXnvtNcNmsxkul6te/Xe5XIYkI2vRFGPrS9PNFwAAaL7OfX/X9/u+IXj0GqjCwkKVlJQoISHBbLPZbOrfv7+2bt0qScrLy1N1dbVbjdPpVHR0tFmTk5Mju92uXr16mTW9e/eW3W53q4mOjpbT6TRrBg8erMrKSuXl5XlyWAAAAG58PLmxkpISSVJERIRbe0REhPbv32/W+Pn5KTg4uFbNufVLSkoUHh5ea/vh4eFuNed/TnBwsPz8/Mya81VWVqqystJ8X15ebmV4AAAAkhroLjwvLy+394Zh1Go73/k1ddX/lJr/lp6ebl6Ubrfb1b59+wv2CQAAoC4eDVAOh0OSas0AlZaWmrNFDodDVVVVKisru2DN4cOHa23/yJEjbjXnf05ZWZmqq6trzUydM3v2bLlcLvN14MCBnzBKAABwpfNogIqMjJTD4VBWVpbZVlVVpezsbPXp00eSFBsbK19fX7ea4uJiFRQUmDVxcXFyuVzavn27WbNt2za5XC63moKCAhUXF5s169evl81mU2xsbJ39s9lsat26tdsLAADAKsvXQB0/flxffvml+b6wsFD5+fkKCQlRhw4dlJaWprlz5yoqKkpRUVGaO3euAgIClJycLEmy2+0aP368pk+frtDQUIWEhGjGjBmKiYnRwIEDJUmdO3fWkCFDNGHCBC1dulSSNHHiRCUmJqpTp06SpISEBHXp0kUpKSl65pln9P3332vGjBmaMGECwQgAADQoywHqo48+0i233GK+nzZtmiRp7NixWrFihWbOnKmKigpNnjxZZWVl6tWrl9avX6+goCBznQULFsjHx0cjR45URUWFBgwYoBUrVsjb29usWb16tVJTU8279ZKSktyePeXt7a333ntPkydPVt++feXv76/k5GTNmzfP+l4AAACwwMswDKOpO9FUysvLZbfblbVoigL9bWZ73HhCGAAAzdW572+Xy9VkZ534LTwAAACLCFAAAAAWEaAAAAAsIkABAABYRIACAACwiAAFAABgEQEKAADAIgIUAACARQQoAAAAiwhQAAAAFhGgAAAALCJAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAAAsIkABAABYRIACAACwiAAFAABgEQEKAADAIgIUAACARQQoAAAAiwhQAAAAFhGgAAAALCJAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIs8HqBOnz6tP/zhD4qMjJS/v7+uvfZaPf744zpz5oxZYxiG5syZI6fTKX9/f8XHx2v37t1u26msrNTUqVMVFhamwMBAJSUl6eDBg241ZWVlSklJkd1ul91uV0pKio4ePerpIQEAALjxeIB66qmn9MILL2jx4sXau3evnn76aT3zzDNatGiRWfP0009r/vz5Wrx4sXbs2CGHw6FBgwbp2LFjZk1aWprWrFmjjIwMbdmyRcePH1diYqJqamrMmuTkZOXn5yszM1OZmZnKz89XSkqKp4cEAADgxsswDMOTG0xMTFRERIRefvlls+1Xv/qVAgICtHLlShmGIafTqbS0NM2aNUvS2dmmiIgIPfXUU5o0aZJcLpfatm2rlStXatSoUZKkb775Ru3bt9e6des0ePBg7d27V126dFFubq569eolScrNzVVcXJw+++wzderU6Uf7Wl5eLrvdrqxFUxTobzPb48bP8+QuAQAAHnTu+9vlcql169ZN0gePz0D169dPH3zwgfbt2ydJ+vjjj7VlyxYNHTpUklRYWKiSkhIlJCSY69hsNvXv319bt26VJOXl5am6utqtxul0Kjo62qzJycmR3W43w5Mk9e7dW3a73aw5X2VlpcrLy91eAAAAVvl4eoOzZs2Sy+XS9ddfL29vb9XU1OiJJ57Q3XffLUkqKSmRJEVERLitFxERof3795s1fn5+Cg4OrlVzbv2SkhKFh4fX+vzw8HCz5nzp6el67LHHLm6AAADgiufxGai//e1vWrVqlV599VXt3LlTr7zyiubNm6dXXnnFrc7Ly8vtvWEYtdrOd35NXfUX2s7s2bPlcrnM14EDB+o7LAAAAJPHZ6AefPBBPfTQQ7rrrrskSTExMdq/f7/S09M1duxYORwOSWdnkNq1a2euV1paas5KORwOVVVVqayszG0WqrS0VH369DFrDh8+XOvzjxw5Umt26xybzSabzVbnMgAAgPry+AzUyZMn1aKF+2a9vb3NxxhERkbK4XAoKyvLXF5VVaXs7GwzHMXGxsrX19etpri4WAUFBWZNXFycXC6Xtm/fbtZs27ZNLpfLrAEAAGgIHp+BGj58uJ544gl16NBBN9xwg3bt2qX58+fr3nvvlXT2tFtaWprmzp2rqKgoRUVFae7cuQoICFBycrIkyW63a/z48Zo+fbpCQ0MVEhKiGTNmKCYmRgMHDpQkde7cWUOGDNGECRO0dOlSSdLEiROVmJhYrzvwAAAAfiqPB6hFixbpkUce0eTJk1VaWiqn06lJkybp0UcfNWtmzpypiooKTZ48WWVlZerVq5fWr1+voKAgs2bBggXy8fHRyJEjVVFRoQEDBmjFihXy9vY2a1avXq3U1FTzbr2kpCQtXrzY00MCAABw4/HnQF1KeA4UAACXnsvyOVAAAACXOwIUAACARQQoAAAAiwhQAAAAFhGgAAAALCJAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAAAsIkABAABYRIACAACwiAAFAABgEQEKAADAIgIUAACARQQoAAAAiwhQAAAAFhGgAAAALCJAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGBRgwSoQ4cO6Z577lFoaKgCAgLUrVs35eXlmcsNw9CcOXPkdDrl7++v+Ph47d69220blZWVmjp1qsLCwhQYGKikpCQdPHjQraasrEwpKSmy2+2y2+1KSUnR0aNHG2JIAAAAJo8HqLKyMvXt21e+vr56//33tWfPHj377LNq06aNWfP0009r/vz5Wrx4sXbs2CGHw6FBgwbp2LFjZk1aWprWrFmjjIwMbdmyRcePH1diYqJqamrMmuTkZOXn5yszM1OZmZnKz89XSkqKp4cEAADgxsswDMOTG3zooYf073//W5s3b65zuWEYcjqdSktL06xZsySdnW2KiIjQU089pUmTJsnlcqlt27ZauXKlRo0aJUn65ptv1L59e61bt06DBw/W3r171aVLF+Xm5qpXr16SpNzcXMXFxemzzz5Tp06dfrSv5eXlstvtylo0RYH+NrM9bvy8i90NAACggZz7/na5XGrdunWT9MHjM1Br165Vz549deeddyo8PFzdu3fXiy++aC4vLCxUSUmJEhISzDabzab+/ftr69atkqS8vDxVV1e71TidTkVHR5s1OTk5stvtZniSpN69e8tut5s156usrFR5ebnbCwAAwCqPB6j//Oc/WrJkiaKiovTPf/5T9913n1JTU/XXv/5VklRSUiJJioiIcFsvIiLCXFZSUiI/Pz8FBwdfsCY8PLzW54eHh5s150tPTzevl7Lb7Wrfvv3FDRYAAFyRPB6gzpw5ox49emju3Lnq3r27Jk2apAkTJmjJkiVudV5eXm7vDcOo1Xa+82vqqr/QdmbPni2Xy2W+Dhw4UN9hAQAAmDweoNq1a6cuXbq4tXXu3FlFRUWSJIfDIUm1ZolKS0vNWSmHw6GqqiqVlZVdsObw4cO1Pv/IkSO1ZrfOsdlsat26tdsLAADAKo8HqL59++rzzz93a9u3b586duwoSYqMjJTD4VBWVpa5vKqqStnZ2erTp48kKTY2Vr6+vm41xcXFKigoMGvi4uLkcrm0fft2s2bbtm1yuVxmDQAAQEPw8fQGf//736tPnz6aO3euRo4cqe3bt2vZsmVatmyZpLOn3dLS0jR37lxFRUUpKipKc+fOVUBAgJKTkyVJdrtd48eP1/Tp0xUaGqqQkBDNmDFDMTExGjhwoKSzs1pDhgzRhAkTtHTpUknSxIkTlZiYWK878AAAAH4qjweom266SWvWrNHs2bP1+OOPKzIyUgsXLtTo0aPNmpkzZ6qiokKTJ09WWVmZevXqpfXr1ysoKMisWbBggXx8fDRy5EhVVFRowIABWrFihby9vc2a1atXKzU11bxbLykpSYsXL/b0kAAAANx4/DlQlxKeAwUAwKXnsnwOFAAAwOWOAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAAAsIkABAABYRIACAACwiAAFAABgEQEKAADAIgIUAACARQQoAAAAiwhQAAAAFhGgAAAALCJAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAAAsIkABAABYRIACAACwiAAFAABgEQEKAADAIgIUAACARQQoAAAAiwhQAAAAFhGgAAAALCJAAQAAWNTgASo9PV1eXl5KS0sz2wzD0Jw5c+R0OuXv76/4+Hjt3r3bbb3KykpNnTpVYWFhCgwMVFJSkg4ePOhWU1ZWppSUFNntdtntdqWkpOjo0aMNPSQAAHCFa9AAtWPHDi1btkxdu3Z1a3/66ac1f/58LV68WDt27JDD4dCgQYN07NgxsyYtLU1r1qxRRkaGtmzZouPHjysxMVE1NTVmTXJysvLz85WZmanMzEzl5+crJSWlIYcEAADQcAHq+PHjGj16tF588UUFBweb7YZhaOHChXr44Yd1xx13KDo6Wq+88opOnjypV199VZLkcrn08ssv69lnn9XAgQPVvXt3rVq1Sp9++qk2bNggSdq7d68yMzP10ksvKS4uTnFxcXrxxRf17rvv6vPPP2+oYQEAADRcgLr//vs1bNgwDRw40K29sLBQJSUlSkhIMNtsNpv69++vrVu3SpLy8vJUXV3tVuN0OhUdHW3W5OTkyG63q1evXmZN7969ZbfbzZrzVVZWqry83O0FAABglU9DbDQjI0M7d+7Ujh07ai0rKSmRJEVERLi1R0REaP/+/WaNn5+f28zVuZpz65eUlCg8PLzW9sPDw82a86Wnp+uxxx6zPiAAAID/4vEZqAMHDuiBBx7QqlWr1LJlyx+s8/LycntvGEattvOdX1NX/YW2M3v2bLlcLvN14MCBC34eAABAXTweoPLy8lRaWqrY2Fj5+PjIx8dH2dnZeu655+Tj42POPJ0/S1RaWmouczgcqqqqUllZ2QVrDh8+XOvzjxw5Umt26xybzabWrVu7vQAAAKzyeIAaMGCAPv30U+Xn55uvnj17avTo0crPz9e1114rh8OhrKwsc52qqiplZ2erT58+kqTY2Fj5+vq61RQXF6ugoMCsiYuLk8vl0vbt282abdu2yeVymTUAAAANwePXQAUFBSk6OtqtLTAwUKGhoWZ7Wlqa5s6dq6ioKEVFRWnu3LkKCAhQcnKyJMlut2v8+PGaPn26QkNDFRISohkzZigmJsa8KL1z584aMmSIJkyYoKVLl0qSJk6cqMTERHXq1MnTwwIAADA1yEXkP2bmzJmqqKjQ5MmTVVZWpl69emn9+vUKCgoyaxYsWCAfHx+NHDlSFRUVGjBggFasWCFvb2+zZvXq1UpNTTXv1ktKStLixYsbfTwAAODK4mUYhtHUnWgq5eXlstvtylo0RYH+NrM9bvy8JuwVAAC4kHPf3y6Xq8muZ+a38AAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAAAsIkABAABYRIACAACwiAAFAABgEQEKAADAIgIUAACARQQoAAAAiwhQAAAAFhGgAAAALCJAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACwiQAEAAFjk09QdaI4WZO2r1fb7QT9rgp4AAIDmiBkoAAAAiwhQAAAAFhGgAAAALCJAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEU8SLOeeLgmAAA4hwB1EQhVAABcmQhQzRwhDQCA5odroAAAACwiQAEAAFjk8QCVnp6um266SUFBQQoPD9eIESP0+eefu9UYhqE5c+bI6XTK399f8fHx2r17t1tNZWWlpk6dqrCwMAUGBiopKUkHDx50qykrK1NKSorsdrvsdrtSUlJ09OhRTw+pQSzI2lfrRV8AALg0eDxAZWdn6/7771dubq6ysrJ0+vRpJSQk6MSJE2bN008/rfnz52vx4sXasWOHHA6HBg0apGPHjpk1aWlpWrNmjTIyMrRlyxYdP35ciYmJqqmpMWuSk5OVn5+vzMxMZWZmKj8/XykpKZ4e0kUjoAAAcHnx+EXkmZmZbu+XL1+u8PBw5eXl6eabb5ZhGFq4cKEefvhh3XHHHZKkV155RREREXr11Vc1adIkuVwuvfzyy1q5cqUGDhwoSVq1apXat2+vDRs2aPDgwdq7d68yMzOVm5urXr16SZJefPFFxcXF6fPPP1enTp08PbRmjYvNAQBoPA1+F57L5ZIkhYSESJIKCwtVUlKihIQEs8Zms6l///7aunWrJk2apLy8PFVXV7vVOJ1ORUdHa+vWrRo8eLBycnJkt9vN8CRJvXv3lt1u19atW+sMUJWVlaqsrDTfl5eXe3y8aBoESABAY2rQAGUYhqZNm6Z+/fopOjpaklRSUiJJioiIcKuNiIjQ/v37zRo/Pz8FBwfXqjm3fklJicLDw2t9Znh4uFlzvvT0dD322GM/2u/eRctqteV2mPij60l1f5FfrpoqtFxJ+xgA0Dw1aICaMmWKPvnkE23ZsqXWMi8vL7f3hmHUajvf+TV11V9oO7Nnz9a0adPM9+Xl5Wrfvv0FP7Mx1TcYXBYB4sP0Wk0LTv+qVtvFBDJmpQAADaXBAtTUqVO1du1abdq0SVdffbXZ7nA4JJ2dQWrXrp3ZXlpaas5KORwOVVVVqayszG0WqrS0VH369DFrDh8+XOtzjxw5Umt26xybzSabzfaTxnMxs1JNhQABAEDD8HiAMgxDU6dO1Zo1a7Rx40ZFRka6LY+MjJTD4VBWVpa6d+8uSaqqqlJ2draeeuopSVJsbKx8fX2VlZWlkSNHSpKKi4tVUFCgp59+WpIUFxcnl8ul7du36+c//7kkadu2bXK5XGbIutTUN6Q15zBX39mx3/MMfADAJczjX2P333+/Xn31Vb399tsKCgoyr0ey2+3y9/eXl5eX0tLSNHfuXEVFRSkqKkpz585VQECAkpOTzdrx48dr+vTpCg0NVUhIiGbMmKGYmBjzrrzOnTtryJAhmjBhgpYuXSpJmjhxohITE5v0DrzmFG7q6os07ydty9OnDXP+813txg51FNZxqq93Ue11672P69henW6ZXb86AMAVyeMBasmSJZKk+Ph4t/bly5dr3LhxkqSZM2eqoqJCkydPVllZmXr16qX169crKCjIrF+wYIF8fHw0cuRIVVRUaMCAAVqxYoW8vb3NmtWrVys1NdW8Wy8pKUmLFy/29JB+UN0BpXl/Rl1ByJNBq7mrK7jFXRtau7CuoOXpUOXpzyAcAkCjaZBTeD/Gy8tLc+bM0Zw5c36wpmXLllq0aJEWLVr0gzUhISFatWrVT+lmo2pOs1L1VZ8Zp8YYV50zVc3dRQSjOq9b83njJ2+vTo0RDnF54m8HMHElyhWmoWfNmiosXsy46j0rVYe6A08dhfWdHbpcXcwXL1/aAJohAlQT8XSQaYzTiQ2tOY2hzlCl+l2PpTrC18WEtLrUfSrWg5/R3ENLc+8fgMseAQoNrjkFo4vh6dOJ9b6Q3sOfUVeoynl5xo/W1NvFzLZ5eqauOQUtD/eFx5QATYsAhTpdLqEHP02d4e4/M2o1xY337M0Gnp6pA4CGQoACPOxiZqrqDK6XWICo7/jjbmngjjSE5jSjdaXjWKCJEaCAK4wnT0Wef+qvsdatUz1P/13Mqa/6XhtX1xd5neteRIisM2x/WEfYbi6hojk9ZoPwBQ8gQAHNXF1fvL115Zxi9fRpvfo+96y+Ac/Twcjj6ggL5/fZ06diPa4x7mL19ONHuB7tskeAAgAPq/MuyboK6xFupOZzHVi9Z+AuQr0fDVLvdet4jtrFqOevI9Q5G3gR6vqx9bpcDsHtUgmkBCgAl5x6X2dV3+Dh4RmO+t6EUd9xXJIPlL0cXC7Pb6vPODiFaRkBCsBli4BSP83p//jreyNFvU9tN5PZOyvqO7Y6H1LcnEPfZXbtGQEKAC4TTXGDQL2fNdZEpyabe/8uhkfv2q0j3NR12rDewfoigtyl8vusBCgAgEd5eubvcpkhvCzGUc/r9urS3AOpVQQoAADqcCnOSnmyz43xXLp633DRDBGgAOAKd6X98sDFzARdFrNIdbhcx9WQCFAAAHhYcwokHr02ztPh08O//9mYCFAAAKBJXMqzny2augMAAACXGgIUAACARQQoAAAAiwhQAAAAFhGgAAAALCJAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEUEKAAAAIsIUAAAABYRoAAAACwiQAEAAFhEgAIAALCIAAUAAGARAQoAAMAiAhQAAIBFBCgAAACLCFAAAAAWEaAAAAAsIkABAABYRIACAACwiAAFAABg0SUfoJ5//nlFRkaqZcuWio2N1ebNm5u6SwAA4DJ3SQeov/3tb0pLS9PDDz+sXbt26Re/+IVuu+02FRUVNXXXAADAZeySDlDz58/X+PHj9dvf/ladO3fWwoUL1b59ey1ZsqSpuwYAAC5jPk3dgZ+qqqpKeXl5euihh9zaExIStHXr1jrXqaysVGVlpfne5XJJkk5UVDVcRwEAwEUpLy+v871hGE3RHUmXcID69ttvVVNTo4iICLf2iIgIlZSU1LlOenq6HnvssVrtI2Yua5A+AgAAD5i6uM7m7777Tna7vZE7c9YlG6DO8fLycntvGEattnNmz56tadOmme+PHj2qjh07qqioqMkOAM4qLy9X+/btdeDAAbVu3bqpu3NF41g0LxyP5oNj0Xy4XC516NBBISEhTdaHSzZAhYWFydvbu9ZsU2lpaa1ZqXNsNptsNlutdrvdzj+GZqJ169Yci2aCY9G8cDyaD45F89GiRdNdyn3JXkTu5+en2NhYZWVlubVnZWWpT58+TdQrAABwJbhkZ6Akadq0aUpJSVHPnj0VFxenZcuWqaioSPfdd19Tdw0AAFzGLukANWrUKH333Xd6/PHHVVxcrOjoaK1bt04dO3as1/o2m01//OMf6zyth8bFsWg+OBbNC8ej+eBYNB/N4Vh4GU15DyAAAMAl6JK9BgoAAKCpEKAAAAAsIkABAABYRIACAACw6IoOUM8//7wiIyPVsmVLxcbGavPmzU3dpUtaenq6brrpJgUFBSk8PFwjRozQ559/7lZjGIbmzJkjp9Mpf39/xcfHa/fu3W41lZWVmjp1qsLCwhQYGKikpCQdPHjQraasrEwpKSmy2+2y2+1KSUnR0aNHG3qIl6T09HR5eXkpLS3NbOM4NK5Dhw7pnnvuUWhoqAICAtStWzfl5eWZyzkejeP06dP6wx/+oMjISPn7++vaa6/V448/rjNnzpg1HIuGsWnTJg0fPlxOp1NeXl5666233JY35n4vKirS8OHDFRgYqLCwMKWmpqqq6if8Jq5xhcrIyDB8fX2NF1980dizZ4/xwAMPGIGBgcb+/fubumuXrMGDBxvLly83CgoKjPz8fGPYsGFGhw4djOPHj5s1Tz75pBEUFGS88cYbxqeffmqMGjXKaNeunVFeXm7W3HfffcZVV11lZGVlGTt37jRuueUW48YbbzROnz5t1gwZMsSIjo42tm7damzdutWIjo42EhMTG3W8l4Lt27cb11xzjdG1a1fjgQceMNs5Do3n+++/Nzp27GiMGzfO2LZtm1FYWGhs2LDB+PLLL80ajkfj+POf/2yEhoYa7777rlFYWGi8/vrrRqtWrYyFCxeaNRyLhrFu3Trj4YcfNt544w1DkrFmzRq35Y2130+fPm1ER0cbt9xyi7Fz504jKyvLcDqdxpQpUyyP6YoNUD//+c+N++67z63t+uuvNx566KEm6tHlp7S01JBkZGdnG4ZhGGfOnDEcDofx5JNPmjWnTp0y7Ha78cILLxiGYRhHjx41fH19jYyMDLPm0KFDRosWLYzMzEzDMAxjz549hiQjNzfXrMnJyTEkGZ999lljDO2ScOzYMSMqKsrIysoy+vfvbwYojkPjmjVrltGvX78fXM7xaDzDhg0z7r33Xre2O+64w7jnnnsMw+BYNJbzA1Rj7vd169YZLVq0MA4dOmTWvPbaa4bNZjNcLpelcVyRp/CqqqqUl5enhIQEt/aEhARt3bq1iXp1+XG5XJJk/thjYWGhSkpK3Pa7zWZT//79zf2el5en6upqtxqn06no6GizJicnR3a7Xb169TJrevfuLbvdzvH7L/fff7+GDRumgQMHurVzHBrX2rVr1bNnT915550KDw9X9+7d9eKLL5rLOR6Np1+/fvrggw+0b98+SdLHH3+sLVu2aOjQoZI4Fk2lMfd7Tk6OoqOj5XQ6zZrBgwersrLS7bR6fVzSTyL/qb799lvV1NTU+tHhiIiIWj9OjJ/GMAxNmzZN/fr1U3R0tCSZ+7au/b5//36zxs/PT8HBwbVqzq1fUlKi8PDwWp8ZHh7O8fv/ZWRkaOfOndqxY0etZRyHxvWf//xHS5Ys0bRp0/T//t//0/bt25WamiqbzaYxY8ZwPBrRrFmz5HK5dP3118vb21s1NTV64okndPfdd0vi30ZTacz9XlJSUutzgoOD5efnZ/nYXJEB6hwvLy+394Zh1GrDTzNlyhR98skn2rJlS61lP2W/n19TVz3H76wDBw7ogQce0Pr169WyZcsfrOM4NI4zZ86oZ8+emjt3riSpe/fu2r17t5YsWaIxY8aYdRyPhve3v/1Nq1at0quvvqobbrhB+fn5SktLk9Pp1NixY806jkXTaKz97qljc0WewgsLC5O3t3ettFlaWlormcK6qVOnau3atfrwww919dVXm+0Oh0OSLrjfHQ6HqqqqVFZWdsGaw4cP1/rcI0eOcPx0dqq7tLRUsbGx8vHxkY+Pj7Kzs/Xcc8/Jx8fH3Ecch8bRrl07denSxa2tc+fOKioqksS/i8b04IMP6qGHHtJdd92lmJgYpaSk6Pe//73S09MlcSyaSmPud4fDUetzysrKVF1dbfnYXJEBys/PT7GxscrKynJrz8rKUp8+fZqoV5c+wzA0ZcoUvfnmm/rXv/6lyMhIt+WRkZFyOBxu+72qqkrZ2dnmfo+NjZWvr69bTXFxsQoKCsyauLg4uVwubd++3azZtm2bXC4Xx0/SgAED9Omnnyo/P9989ezZU6NHj1Z+fr6uvfZajkMj6tu3b63Heezbt8/80XP+XTSekydPqkUL9689b29v8zEGHIum0Zj7PS4uTgUFBSouLjZr1q9fL5vNptjYWGsdt3TJ+WXk3GMMXn75ZWPPnj1GWlqaERgYaHz99ddN3bVL1u9+9zvDbrcbGzduNIqLi83XyZMnzZonn3zSsNvtxptvvml8+umnxt13313nrapXX321sWHDBmPnzp3GrbfeWuetql27djVycnKMnJwcIyYm5oq+RfjH/PddeIbBcWhM27dvN3x8fIwnnnjC+OKLL4zVq1cbAQEBxqpVq8wajkfjGDt2rHHVVVeZjzF48803jbCwMGPmzJlmDceiYRw7dszYtWuXsWvXLkOSMX/+fGPXrl3mo4Maa7+fe4zBgAEDjJ07dxobNmwwrr76ah5jYNX//d//GR07djT8/PyMHj16mLfb46eRVOdr+fLlZs2ZM2eMP/7xj4bD4TBsNptx8803G59++qnbdioqKowpU6YYISEhhr+/v5GYmGgUFRW51Xz33XfG6NGjjaCgICMoKMgYPXq0UVZW1gijvDSdH6A4Do3rnXfeMaKjow2bzWZcf/31xrJly9yWczwaR3l5ufHAAw8YHTp0MFq2bGlce+21xsMPP2xUVlaaNRyLhvHhhx/W+f0wduxYwzAad7/v37/fGDZsmOHv72+EhIQYU6ZMMU6dOmV5TF6GYRjW5qwAAACubFfkNVAAAAAXgwAFAABgEQEKAADAIgIUAACARQQoAAAAiwhQAAAAFhGgAAAALCJAAQAAWESAAgAAsIgABQAAYBEBCgAAwCICFAAAgEX/H6QoLlQ4/W7sAAAAAElFTkSuQmCC\n",
+ "text/plain": [
+ "<Figure size 640x480 with 1 Axes>"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ }
+ ],
+ "source": [
+ "plt.hist( peaks_closest_upstream_dir['distance'], np.arange(0, 1e4, 100), alpha=0.5, label=\"upstream\");\n",
+ "plt.hist( peaks_closest_downstream_dir['distance'], np.arange(0, 1e4, 100), alpha=0.5, label=\"downstream\");\n",
+ "plt.xlim( [0, 1e4] )\n",
+ "plt.legend()"
+ ]
+ },
+ {
+ "cell_type": "markdown",
+ "id": "65b4a264",
+ "metadata": {},
+ "source": [
+ "CTCF peaks upstream of the genes are more enriched at short distances to TSS, if we take the strand into account."
+ ]
+ }
+ ],
+ "metadata": {
+ "kernelspec": {
+ "display_name": "Python 3",
+ "language": "python",
+ "name": "python3"
+ },
+ "language_info": {
+ "codemirror_mode": {
+ "name": "ipython",
+ "version": 3
+ },
+ "file_extension": ".py",
+ "mimetype": "text/x-python",
+ "name": "python",
+ "nbconvert_exporter": "python",
+ "pygments_lexer": "ipython3",
+ "version": "3.10.4"
+ }
+ },
+ "nbformat": 4,
+ "nbformat_minor": 5
+}
=====================================
tests/test_core_specs.py
=====================================
@@ -69,7 +69,7 @@ def test_verify_columns():
# no repeated column names
with pytest.raises(ValueError):
- specs._verify_columns(df1, ["chromStart", "chromStart"])
+ specs._verify_columns(df1, ["chromStart", "chromStart"], unique_cols=True)
def test_verify_column_dtypes():
=====================================
tests/test_extras.py
=====================================
@@ -178,6 +178,19 @@ def test_frac_gc():
).all()
+def test_seq_gc():
+
+ assert (0 == bioframe.seq_gc("AT"))
+ assert (np.isnan( bioframe.seq_gc("NNN")))
+ assert (1 == bioframe.seq_gc("NGnC"))
+ assert (0.5 == bioframe.seq_gc("GTCA"))
+ assert (0.25 == bioframe.seq_gc("nnnNgTCa", mapped_only=False))
+ with pytest.raises(ValueError):
+ bioframe.seq_gc(["A", "T"])
+ with pytest.raises(ValueError):
+ bioframe.seq_gc(np.array("ATGC"))
+
+
### todo: test frac_gene_coverage(bintable, mrna):
### currently broken
=====================================
tests/test_ops.py
=====================================
@@ -64,74 +64,6 @@ def mock_bioframe(num_entries=100):
############# tests #####################
-def test_select():
- df1 = pd.DataFrame(
- [["chrX", 3, 8], ["chr1", 4, 5], ["chrX", 1, 5]],
- columns=["chrom", "start", "end"],
- )
-
- region1 = "chr1:4-10"
- df_result = pd.DataFrame([["chr1", 4, 5]], columns=["chrom", "start", "end"])
- pd.testing.assert_frame_equal(
- df_result, bioframe.select(df1, region1).reset_index(drop=True)
- )
-
- region1 = "chrX"
- df_result = pd.DataFrame(
- [["chrX", 3, 8], ["chrX", 1, 5]], columns=["chrom", "start", "end"]
- )
- pd.testing.assert_frame_equal(
- df_result, bioframe.select(df1, region1).reset_index(drop=True)
- )
-
- region1 = "chrX:4-6"
- df_result = pd.DataFrame(
- [["chrX", 3, 8], ["chrX", 1, 5]], columns=["chrom", "start", "end"]
- )
- pd.testing.assert_frame_equal(
- df_result, bioframe.select(df1, region1).reset_index(drop=True)
- )
-
- ### select with non-standard column names
- region1 = "chrX:4-6"
- new_names = ["chr", "chrstart", "chrend"]
- df1 = pd.DataFrame(
- [["chrX", 3, 8], ["chr1", 4, 5], ["chrX", 1, 5]],
- columns=new_names,
- )
- df_result = pd.DataFrame(
- [["chrX", 3, 8], ["chrX", 1, 5]],
- columns=new_names,
- )
- pd.testing.assert_frame_equal(
- df_result, bioframe.select(df1, region1, cols=new_names).reset_index(drop=True)
- )
- region1 = "chrX"
- pd.testing.assert_frame_equal(
- df_result, bioframe.select(df1, region1, cols=new_names).reset_index(drop=True)
- )
-
- ### select from a DataFrame with NaNs
- colnames = ["chrom", "start", "end", "view_region"]
- df = pd.DataFrame(
- [
- ["chr1", -6, 12, "chr1p"],
- [pd.NA, pd.NA, pd.NA, "chr1q"],
- ["chrX", 1, 8, "chrX_0"],
- ],
- columns=colnames,
- ).astype({"start": pd.Int64Dtype(), "end": pd.Int64Dtype()})
- df_result = pd.DataFrame(
- [["chr1", -6, 12, "chr1p"]],
- columns=colnames,
- ).astype({"start": pd.Int64Dtype(), "end": pd.Int64Dtype()})
-
- region1 = "chr1:0-1"
- pd.testing.assert_frame_equal(
- df_result, bioframe.select(df, region1).reset_index(drop=True)
- )
-
-
def test_trim():
### trim with view_df
@@ -931,19 +863,6 @@ def test_closest():
df, bioframe.closest(df1, df2, suffixes=("_1", "_2"), k=2)
)
- ### closest(df2,df1) ###
- d = """chrom_1 start_1 end_1 chrom_2 start_2 end_2 distance
- 0 chr1 4 8 chr1 1 5 0
- 1 chr1 10 11 chr1 1 5 5 """
- df = pd.read_csv(StringIO(d), sep=r"\s+").astype(
- {
- "start_2": pd.Int64Dtype(),
- "end_2": pd.Int64Dtype(),
- "distance": pd.Int64Dtype(),
- }
- )
- pd.testing.assert_frame_equal(df, bioframe.closest(df2, df1, suffixes=("_1", "_2")))
-
### change first interval to new chrom ###
df2.iloc[0, 0] = "chrA"
d = """chrom start end chrom_ start_ end_ distance
@@ -1039,6 +958,74 @@ def test_closest():
df1.iloc[0, 0] = "chr10"
bioframe.closest(df1, df2)
+ ### closest with direction ###
+
+ df1 = pd.DataFrame(
+ [
+ ["chr1", 3, 5, "+"],
+ ["chr1", 3, 5, "-"],
+ ],
+ columns=["chrom", "start", "end", "strand"],
+ )
+
+ df2 = pd.DataFrame(
+ [["chr1", 1, 2], ["chr1", 2, 8], ["chr1", 10, 11]], columns=["chrom", "start", "end"]
+ )
+
+ ### closest(df1, df2, k=1, direction_col="strand") ###
+ d = """chrom start end strand chrom_ start_ end_ distance
+ 0 chr1 3 5 + chr1 2 8 0
+ 1 chr1 3 5 - chr1 2 8 0
+ """
+ df = pd.read_csv(StringIO(d), sep=r"\s+").astype(
+ {
+ "start_": pd.Int64Dtype(),
+ "end_": pd.Int64Dtype(),
+ "distance": pd.Int64Dtype(),
+ }
+ )
+ pd.testing.assert_frame_equal(df, bioframe.closest(df1, df2, k=1, direction_col="strand"))
+
+ ### closest(df1, df2, k=1, ignore_upstream=False, ignore_downstream=True, ignore_overlaps=True, direction_col="strand") ###
+ d = """chrom start end strand chrom_ start_ end_ distance
+ 0 chr1 3 5 + chr1 1 2 1
+ 1 chr1 3 5 - chr1 10 11 5
+ """
+ df = pd.read_csv(StringIO(d), sep=r"\s+").astype(
+ {
+ "start_": pd.Int64Dtype(),
+ "end_": pd.Int64Dtype(),
+ "distance": pd.Int64Dtype(),
+ }
+ )
+ pd.testing.assert_frame_equal(df,
+ bioframe.closest(df1, df2,
+ k=1,
+ ignore_upstream=False,
+ ignore_downstream=True,
+ ignore_overlaps=True,
+ direction_col="strand"))
+
+ ### closest(df1, df2, k=1, ignore_upstream=True, ignore_downstream=False, ignore_overlaps=True, direction_col="strand") ###
+ d = """chrom start end strand chrom_ start_ end_ distance
+ 0 chr1 3 5 + chr1 10 11 5
+ 1 chr1 3 5 - chr1 1 2 1
+ """
+ df = pd.read_csv(StringIO(d), sep=r"\s+").astype(
+ {
+ "start_": pd.Int64Dtype(),
+ "end_": pd.Int64Dtype(),
+ "distance": pd.Int64Dtype(),
+ }
+ )
+ pd.testing.assert_frame_equal(df,
+ bioframe.closest(df1, df2,
+ k=1,
+ ignore_upstream=True,
+ ignore_downstream=False,
+ ignore_overlaps=True,
+ direction_col="strand"))
+
def test_coverage():
=====================================
tests/test_ops_select.py
=====================================
@@ -0,0 +1,230 @@
+import pandas as pd
+import numpy as np
+import pytest
+
+import bioframe
+
+
+def test_select():
+ df = pd.DataFrame(
+ [["chrX", 3, 8],
+ ["chr1", 4, 5],
+ ["chrX", 1, 5]],
+ columns=["chrom", "start", "end"],
+ )
+
+ result = pd.DataFrame(
+ [["chr1", 4, 5]],
+ columns=["chrom", "start", "end"]
+ )
+ pd.testing.assert_frame_equal(
+ result, bioframe.select(df, "chr1:4-10").reset_index(drop=True)
+ )
+
+ result = pd.DataFrame(
+ [["chrX", 3, 8],
+ ["chrX", 1, 5]],
+ columns=["chrom", "start", "end"]
+ )
+ pd.testing.assert_frame_equal(
+ result, bioframe.select(df, "chrX").reset_index(drop=True)
+ )
+
+ result = pd.DataFrame(
+ [["chrX", 3, 8],
+ ["chrX", 1, 5]],
+ columns=["chrom", "start", "end"]
+ )
+ pd.testing.assert_frame_equal(
+ result, bioframe.select(df, "chrX:4-6").reset_index(drop=True)
+ )
+
+ # Query range not in the dataframe
+ assert len(bioframe.select(df, "chrZ")) == 0
+ assert len(bioframe.select(df, "chr1:100-1000")) == 0
+ assert len(bioframe.select(df, "chr1:1-3")) == 0
+
+ # Invalid query range
+ with pytest.raises(ValueError):
+ bioframe.select(df, "chr1:1-0")
+
+
+def test_select__with_colnames():
+ ### select with non-standard column names
+ new_names = ["chr", "chrstart", "chrend"]
+ df = pd.DataFrame(
+ [["chrX", 3, 8],
+ ["chr1", 4, 5],
+ ["chrX", 1, 5]],
+ columns=new_names,
+ )
+ result = pd.DataFrame(
+ [["chrX", 3, 8],
+ ["chrX", 1, 5]],
+ columns=new_names,
+ )
+ pd.testing.assert_frame_equal(
+ result, bioframe.select(df, "chrX:4-6", cols=new_names).reset_index(drop=True)
+ )
+ pd.testing.assert_frame_equal(
+ result, bioframe.select(df, "chrX", cols=new_names).reset_index(drop=True)
+ )
+
+
+def test_select__with_nulls():
+ ### select from a DataFrame with NaNs
+ colnames = ["chrom", "start", "end", "view_region"]
+ df = pd.DataFrame(
+ [
+ ["chr1", -6, 12, "chr1p"],
+ [pd.NA, pd.NA, pd.NA, "chr1q"],
+ ["chrX", 1, 8, "chrX_0"],
+ ],
+ columns=colnames,
+ ).astype({"start": pd.Int64Dtype(), "end": pd.Int64Dtype()})
+
+ result = pd.DataFrame(
+ [["chr1", -6, 12, "chr1p"]],
+ columns=colnames,
+ ).astype({"start": pd.Int64Dtype(), "end": pd.Int64Dtype()})
+
+ pd.testing.assert_frame_equal(
+ result, bioframe.select(df, "chr1:0-1").reset_index(drop=True)
+ )
+
+
+def test_select__mask_indices_labels():
+ df = pd.DataFrame(
+ [["chrX", 3, 8],
+ ["chr1", 4, 5],
+ ["chrX", 1, 5]],
+ columns=["chrom", "start", "end"],
+ )
+
+ region = "chr1:4-10"
+ answer = pd.DataFrame(
+ [["chr1", 4, 5]],
+ columns=["chrom", "start", "end"]
+ )
+
+ result = bioframe.select(df, region)
+ pd.testing.assert_frame_equal(
+ answer, result.reset_index(drop=True)
+ )
+ mask = bioframe.select_mask(df, region)
+ pd.testing.assert_frame_equal(
+ answer, df.loc[mask].reset_index(drop=True)
+ )
+ labels = bioframe.select_labels(df, region)
+ pd.testing.assert_frame_equal(
+ answer, df.loc[labels].reset_index(drop=True)
+ )
+ idx = bioframe.select_indices(df, region)
+ pd.testing.assert_frame_equal(
+ answer, df.iloc[idx].reset_index(drop=True)
+ )
+
+
+def test_select__query_intervals_are_half_open():
+ df = pd.DataFrame({
+ "chrom": ["chr1", "chr1",
+ "chr2", "chr2", "chr2", "chr2", "chr2", "chr2"],
+ "start": [0, 10,
+ 10, 20, 30, 40, 50, 60],
+ "end": [10, 20,
+ 20, 30, 40, 50, 60, 70],
+ "name": ["a", "b",
+ "A", "B", "C", "D", "E", "F"],
+ })
+
+ result = bioframe.select(df, "chr1")
+ assert (result["name"] == ["a", "b"]).all()
+
+ result = bioframe.select(df, "chr2:20-70")
+ assert (result["name"] == ["B", "C", "D", "E", "F"]).all()
+
+ result = bioframe.select(df, "chr2:20-75")
+ assert (result["name"] == ["B", "C", "D", "E", "F"]).all()
+
+ result = bioframe.select(df, "chr2:20-")
+ assert (result.index == [3, 4, 5, 6, 7]).all()
+
+ result = bioframe.select(df, "chr2:20-30")
+ assert (result["name"] == ["B"]).all()
+
+ result = bioframe.select(df, "chr2:20-40")
+ assert (result["name"] == ["B", "C"]).all()
+
+ result = bioframe.select(df, "chr2:20-45")
+ assert (result["name"] == ["B", "C", "D"]).all()
+
+ result = bioframe.select(df, "chr2:19-45")
+ assert (result["name"] == ["A", "B", "C", "D"]).all()
+
+ result = bioframe.select(df, "chr2:25-45")
+ assert (result["name"] == ["B", "C", "D"]).all()
+
+ result = bioframe.select(df, "chr2:25-50")
+ assert (result["name"] == ["B", "C", "D"]).all()
+
+ result = bioframe.select(df, "chr2:25-51")
+ assert (result["name"] == ["B", "C", "D", "E"]).all()
+
+
+def test_select__with_point_intervals():
+ # Dataframe containing "point intervals"
+ df = pd.DataFrame({
+ "chrom": ["chr1", "chr1",
+ "chr2", "chr2", "chr2", "chr2", "chr2", "chr2"],
+ "start": [0, 10,
+ 10, 20, 30, 40, 50, 60],
+ "end": [10, 10,
+ 20, 30, 40, 50, 50, 70],
+ "name": ["a", "b",
+ "A", "B", "C", "D", "E", "F"],
+ })
+ result = bioframe.select(df, "chr1")
+ assert (result["name"] == ["a", "b"]).all()
+
+ result = bioframe.select(df, "chr1:4-10")
+ assert (result["name"] == ["a"]).all()
+
+ result = bioframe.select(df, "chr1:4-4")
+ assert (result["name"] == ["a"]).all()
+
+ result = bioframe.select(df, "chr1:10-15")
+ assert (result["name"] == ["b"]).all()
+
+ result = bioframe.select(df, "chr2:20-70")
+ assert (result["name"] == ["B", "C", "D", "E", "F"]).all()
+
+ result = bioframe.select(df, "chr2:49-70")
+ assert (result["name"] == ["D", "E", "F"]).all()
+
+ result = bioframe.select(df, "chr2:50-70")
+ assert (result["name"] == ["E", "F"]).all()
+
+ result = bioframe.select(df, "chr2:50-51")
+ assert (result["name"] == ["E"]).all()
+
+ result = bioframe.select(df, "chr2:50-50")
+ assert (result["name"] == ["E"]).all()
+
+
+def test_select__with_points():
+ # Dataframe of points
+ df = pd.DataFrame(
+ [["chrX", 3, "A"],
+ ["chr1", 4, "C"],
+ ["chrX", 1, "B"]],
+ columns=["chrom", "pos", "name"],
+ )
+
+ result = bioframe.select(df, "chr1:4-10", cols=["chrom", "pos", "pos"])
+ assert (result["name"] == ["C"]).all()
+
+ result = bioframe.select(df, "chr1:3-10", cols=["chrom", "pos", "pos"])
+ assert (result["name"] == ["C"]).all()
+
+ result = bioframe.select(df, "chr1:4-4", cols=["chrom", "pos", "pos"])
+ assert (result["name"] == ["C"]).all()
View it on GitLab: https://salsa.debian.org/med-team/python-bioframe/-/commit/0476c7c37de726aa62e88b74c7be210cd83bed16
--
View it on GitLab: https://salsa.debian.org/med-team/python-bioframe/-/commit/0476c7c37de726aa62e88b74c7be210cd83bed16
You're receiving this email because of your account on salsa.debian.org.
-------------- next part --------------
An HTML attachment was scrubbed...
URL: <http://alioth-lists.debian.net/pipermail/debian-med-commit/attachments/20230507/91f280c7/attachment-0001.htm>
More information about the debian-med-commit
mailing list