[med-svn] [Git][med-team/unanimity][master] 6 commits: Standards-Version: 4.4.1
Andreas Tille
gitlab at salsa.debian.org
Thu Dec 19 08:00:15 GMT 2019
Andreas Tille pushed to branch master at Debian Med / unanimity
Commits:
ee41f701 by Andreas Tille at 2019-12-19T07:09:24Z
Standards-Version: 4.4.1
- - - - -
ae9de4ef by Andreas Tille at 2019-12-19T07:11:54Z
Update patch
- - - - -
60fdd657 by Andreas Tille at 2019-12-19T07:15:01Z
Use 2to3 to port from Python2 to Python3
- - - - -
f48ff806 by Andreas Tille at 2019-12-19T07:16:55Z
Switch to Python3 in packaging
- - - - -
5adf3be5 by Andreas Tille at 2019-12-19T07:17:31Z
debian/copyright: use spaces rather than tabs to start continuation lines.
Fixes lintian: tab-in-licence-text
See https://lintian.debian.org/tags/tab-in-licence-text.html for more details.
- - - - -
07c8d23d by Andreas Tille at 2019-12-19T07:59:46Z
Close proper bugs
- - - - -
7 changed files:
- debian/changelog
- debian/control
- debian/copyright
- + debian/patches/2to3.patch
- debian/patches/do_not_refer_to_non-existing_images.patch
- debian/patches/series
- debian/rules
Changes:
=====================================
debian/changelog
=====================================
@@ -4,10 +4,15 @@ unanimity (3.4.1+git20180307.02aa264+dfsg-1) UNRELEASED; urgency=medium
* New upstream version
Attention: This is the last release of unanimity obtained from Git
(see README.source)
+ Closes: #939506
* debhelper-compat 12
- * Standards-Version: 4.4.0
+ * Standards-Version: 4.4.1
+ * Use 2to3 to port from Python2 to Python3
+ Closes: #938751
+ * debian/copyright: use spaces rather than tabs to start continuation
+ lines.
- -- Andreas Tille <tille at debian.org> Tue, 17 Sep 2019 08:03:35 +0200
+ -- Andreas Tille <tille at debian.org> Thu, 19 Dec 2019 08:17:37 +0100
unanimity (3.3.0+dfsg-2.1) unstable; urgency=medium
=====================================
debian/control
=====================================
@@ -8,16 +8,16 @@ Build-Depends: debhelper-compat (= 12),
dh-python,
cmake,
swig,
- python-setuptools,
- python-all-dev,
- python-numpy,
+ python3-setuptools,
+ python3-all-dev,
+ python3-numpy,
libboost-dev,
libhts-dev,
libpbbam-dev (>= 0.18.0+dfsg-1~),
libpbcopper-dev,
libseqan2-dev,
pandoc
-Standards-Version: 4.4.0
+Standards-Version: 4.4.1
Vcs-Browser: https://salsa.debian.org/med-team/unanimity
Vcs-Git: https://salsa.debian.org/med-team/unanimity.git
Homepage: https://github.com/PacificBiosciences/unanimity
@@ -26,7 +26,7 @@ Package: unanimity
Architecture: any
Depends: ${shlibs:Depends},
${misc:Depends},
- ${python:Depends}
+ ${python3:Depends}
Description: generate and process accurate consensus nucleotide sequences
Unanimity provides a set of tools for consensus sequences from Pacific
Biosciences sequencing data:
@@ -37,17 +37,17 @@ Description: generate and process accurate consensus nucleotide sequences
* Minor variant caller
juliet identifies minor variants from aligned ccs reads.
-Package: python-consensuscore2
+Package: python3-consensuscore2
Architecture: any
Section: python
Depends: ${shlibs:Depends},
${misc:Depends},
- ${python:Depends}
-Description: generate consensus sequences for PacBio data -- Python 2
+ ${python3:Depends}
+Description: generate consensus sequences for PacBio data -- Python3
ConsensusCore2 embodies core C++ routines underlying the Arrow HMM
algorithm for PacBio multi-sequence consensus. Arrow is the successor
to the Quiver model---a CRF model that was embodied in the
ConsensusCore C++ library. Compared to Quiver, the Arrow model is more
statistically principled and easier and more robust to train.
.
- This package installs the library for Python 2.
+ This package installs the library for Python3.
=====================================
debian/copyright
=====================================
@@ -3,15 +3,15 @@ Upstream-Name: unanimity
Upstream-Contact: Pacific Biosciences <devnet at pacb.com>
Source: https://github.com/PacificBiosciences/unanimity
Files-Excluded:
- tools/Darwin
- tools/Linux
- tools/win32
- third-party/cram-*
+ tools/Darwin
+ tools/Linux
+ tools/win32
+ third-party/cram-*
Files: *
Copyright:
- 2011-2016 Pacific Biosciences of California
- 2016 David Alexander, Nigel Delaney, Lance Hepler, Armin Töpfer
+ 2011-2016 Pacific Biosciences of California
+ 2016 David Alexander, Nigel Delaney, Lance Hepler, Armin Töpfer
License: BSD-3-Clause-PacBio
Files: cmake/uny-codecoverage.cmake
=====================================
debian/patches/2to3.patch
=====================================
@@ -0,0 +1,257 @@
+Description: Use 2to3 to port from Python2 to Python3
+Bug-Debian: https://bugs.debian.org/938751
+Author: Andreas Tille <tille at debian.org>
+Last-Update: Thu, 19 Dec 2019 08:14:41 +0100
+
+--- a/doc/ConsensusCore2-DesignAndImplementation/conf.py
++++ b/doc/ConsensusCore2-DesignAndImplementation/conf.py
+@@ -51,18 +51,18 @@ source_suffix = '.rst'
+ master_doc = 'index'
+
+ # General information about the project.
+-project = u'ConsensusCore2: Design and Implementation'
+-copyright = u'2016, David Alexander, Nigel Delaney, Lance Hepler, Armin Töpfer'
+-author = u'David Alexander, Nigel Delaney, Lance Hepler, Armin Töpfer'
++project = 'ConsensusCore2: Design and Implementation'
++copyright = '2016, David Alexander, Nigel Delaney, Lance Hepler, Armin Töpfer'
++author = 'David Alexander, Nigel Delaney, Lance Hepler, Armin Töpfer'
+
+ # The version info for the project you're documenting, acts as replacement for
+ # |version| and |release|, also used in various other places throughout the
+ # built documents.
+ #
+ # The short X.Y version.
+-version = u'0.1'
++version = '0.1'
+ # The full version, including alpha/beta/rc tags.
+-release = u'0.1'
++release = '0.1'
+
+ # The language for content autogenerated by Sphinx. Refer to documentation
+ # for a list of supported languages.
+@@ -262,8 +262,8 @@ latex_elements = {
+ # (source start file, target name, title,
+ # author, documentclass [howto, manual, or own class]).
+ latex_documents = [
+- (master_doc, 'ConsensusCore2DesignandImplementation.tex', u'ConsensusCore2: Design and Implementation Documentation',
+- u'David Alexander, Nigel Delaney, Lance Hepler, Armin Töpfer', 'manual'),
++ (master_doc, 'ConsensusCore2DesignandImplementation.tex', 'ConsensusCore2: Design and Implementation Documentation',
++ 'David Alexander, Nigel Delaney, Lance Hepler, Armin Töpfer', 'manual'),
+ ]
+
+ # The name of an image file (relative to this directory) to place at the top of
+@@ -298,7 +298,7 @@ latex_documents = [
+ # One entry per manual page. List of tuples
+ # (source start file, name, description, authors, manual section).
+ man_pages = [
+- (master_doc, 'consensuscore2designandimplementation', u'ConsensusCore2: Design and Implementation Documentation',
++ (master_doc, 'consensuscore2designandimplementation', 'ConsensusCore2: Design and Implementation Documentation',
+ [author], 1)
+ ]
+
+@@ -313,7 +313,7 @@ man_pages = [
+ # (source start file, target name, title, author,
+ # dir menu entry, description, category)
+ texinfo_documents = [
+- (master_doc, 'ConsensusCore2DesignandImplementation', u'ConsensusCore2: Design and Implementation Documentation',
++ (master_doc, 'ConsensusCore2DesignandImplementation', 'ConsensusCore2: Design and Implementation Documentation',
+ author, 'ConsensusCore2DesignandImplementation', 'One line description of project.',
+ 'Miscellaneous'),
+ ]
+--- a/meson.build
++++ b/meson.build
+@@ -89,7 +89,7 @@ subdir('src')
+ ########
+
+ if (not meson.is_subproject()) and get_option('swig')
+- uny_python_interp = find_program(get_option('PYTHON'), required : true)
++ uny_python_interp = find_program(get_option('PYTHON3'), required : true)
+ uny_swig = find_program('swig', required : true)
+
+ uny_pv_ver = run_command(uny_python_interp, ['-c', 'import sys; sys.stdout.write(sys.version[:3])']).stdout()
+@@ -114,8 +114,8 @@ if (not meson.is_subproject()) and get_o
+
+ message('Python modules will be installed into: ' + uny_python_install_dir)
+
+- # python2 compiling + linking
+- uny_python_dep = dependency('python-2.7', required : true)
++ # python3 compiling + linking
++ uny_python_dep = dependency('python3', required : true)
+
+ subdir('swig')
+ endif
+--- a/scripts/ci/build.sh
++++ b/scripts/ci/build.sh
+@@ -97,11 +97,11 @@ LDFLAGS='-static-libstdc++' \
+ CMAKE_BUILD_TYPE=ReleaseWithAssert \
+ CMAKE_COMMAND=cmake \
+ VERBOSE=1 \
+-$PIP install --user --no-index --verbose .
+-ldd unyve/lib/python2.7/site-packages/_ConsensusCore2.so
++$PIP3 install --user --no-index --verbose .
++ldd unyve/lib/python3.7/site-packages/_ConsensusCore2.so
+
+ echo "## CC2 version test"
+-python -c "import ConsensusCore2 ; print ConsensusCore2.__version__"
++python3 -c "import ConsensusCore2 ; print ConsensusCore2.__version__"
+
+ rm -f unyve/bin/g++
+ module unload ccache
+@@ -114,13 +114,13 @@ module load ccache
+ mv g++ unyve/bin/
+ echo "## Install ConsensusCore"
+ ( cd _deps/ConsensusCore \
+- && python setup.py \
++ && python3 setup.py \
+ bdist_wheel \
+ --boost=$BOOST_ROOT \
+ && echo dist/ConsensusCore-*.whl | \
+ xargs $PIP install --user --verbose )
+ rm unyve/bin/g++
+-ldd unyve/lib/python2.7/site-packages/_ConsensusCore.so
++ldd unyve/lib/python3.7/site-packages/_ConsensusCore.so
+
+ echo "## Install GC"
+ $PIP install --user --no-index --verbose _rev_deps/GenomicConsensus
+--- a/setup.py
++++ b/setup.py
+@@ -1,5 +1,5 @@
+
+-from __future__ import print_function
++
+
+ import os
+ import os.path
+@@ -15,8 +15,8 @@ from subprocess import Popen
+
+ def iteritems(d):
+ if sys.version_info >= (3, 0):
+- return d.items()
+- return d.iteritems()
++ return list(d.items())
++ return iter(d.items())
+
+ def ParseVersion():
+ thisDir = os.path.dirname(os.path.realpath(__file__))
+--- a/tests/python/test_tool_contract.py
++++ b/tests/python/test_tool_contract.py
+@@ -1,4 +1,4 @@
+-#!/usr/bin/env python
++#!/usr/bin/python3
+
+ """
+ Test for tool contract interface support, using a SubreadSet with filters
+--- a/tools/git-clang-format
++++ b/tools/git-clang-format
+@@ -1,4 +1,4 @@
+-#!/usr/bin/env python
++#!/usr/bin/python3
+ #
+ #===- git-clang-format - ClangFormat Git Integration ---------*- python -*--===#
+ #
+@@ -128,15 +128,15 @@ def main():
+ if opts.verbose >= 1:
+ ignored_files.difference_update(changed_lines)
+ if ignored_files:
+- print 'Ignoring changes in the following files (wrong extension):'
++ print('Ignoring changes in the following files (wrong extension):')
+ for filename in ignored_files:
+- print ' ', filename
++ print(' ', filename)
+ if changed_lines:
+- print 'Running clang-format on the following files:'
++ print('Running clang-format on the following files:')
+ for filename in changed_lines:
+- print ' ', filename
++ print(' ', filename)
+ if not changed_lines:
+- print 'no modified files to format'
++ print('no modified files to format')
+ return
+ # The computed diff outputs absolute paths, so we must cd before accessing
+ # those files.
+@@ -146,20 +146,20 @@ def main():
+ binary=opts.binary,
+ style=opts.style)
+ if opts.verbose >= 1:
+- print 'old tree:', old_tree
+- print 'new tree:', new_tree
++ print('old tree:', old_tree)
++ print('new tree:', new_tree)
+ if old_tree == new_tree:
+ if opts.verbose >= 0:
+- print 'clang-format did not modify any files'
++ print('clang-format did not modify any files')
+ elif opts.diff:
+ print_diff(old_tree, new_tree)
+ else:
+ changed_files = apply_changes(old_tree, new_tree, force=opts.force,
+ patch_mode=opts.patch)
+ if (opts.verbose >= 0 and not opts.patch) or opts.verbose >= 1:
+- print 'changed files:'
++ print('changed files:')
+ for filename in changed_files:
+- print ' ', filename
++ print(' ', filename)
+
+
+ def load_git_config(non_string_options=None):
+@@ -298,7 +298,7 @@ def filter_by_extension(dictionary, allo
+ `allowed_extensions` must be a collection of lowercase file extensions,
+ excluding the period."""
+ allowed_extensions = frozenset(allowed_extensions)
+- for filename in dictionary.keys():
++ for filename in list(dictionary.keys()):
+ base_ext = filename.rsplit('.', 1)
+ if len(base_ext) == 1 or base_ext[1].lower() not in allowed_extensions:
+ del dictionary[filename]
+@@ -323,7 +323,7 @@ def run_clang_format_and_save_to_tree(ch
+
+ Returns the object ID (SHA-1) of the created tree."""
+ def index_info_generator():
+- for filename, line_ranges in changed_lines.iteritems():
++ for filename, line_ranges in changed_lines.items():
+ mode = oct(os.stat(filename).st_mode)
+ blob_id = clang_format_to_blob(filename, line_ranges, binary=binary,
+ style=style)
+@@ -431,10 +431,10 @@ def apply_changes(old_tree, new_tree, fo
+ if not force:
+ unstaged_files = run('git', 'diff-files', '--name-status', *changed_files)
+ if unstaged_files:
+- print >>sys.stderr, ('The following files would be modified but '
+- 'have unstaged changes:')
+- print >>sys.stderr, unstaged_files
+- print >>sys.stderr, 'Please commit, stage, or stash them first.'
++ print(('The following files would be modified but '
++ 'have unstaged changes:'), file=sys.stderr)
++ print(unstaged_files, file=sys.stderr)
++ print('Please commit, stage, or stash them first.', file=sys.stderr)
+ sys.exit(2)
+ if patch_mode:
+ # In patch mode, we could just as well create an index from the new tree
+@@ -464,20 +464,20 @@ def run(*args, **kwargs):
+ if p.returncode == 0:
+ if stderr:
+ if verbose:
+- print >>sys.stderr, '`%s` printed to stderr:' % ' '.join(args)
+- print >>sys.stderr, stderr.rstrip()
++ print('`%s` printed to stderr:' % ' '.join(args), file=sys.stderr)
++ print(stderr.rstrip(), file=sys.stderr)
+ if strip:
+ stdout = stdout.rstrip('\r\n')
+ return stdout
+ if verbose:
+- print >>sys.stderr, '`%s` returned %s' % (' '.join(args), p.returncode)
++ print('`%s` returned %s' % (' '.join(args), p.returncode), file=sys.stderr)
+ if stderr:
+- print >>sys.stderr, stderr.rstrip()
++ print(stderr.rstrip(), file=sys.stderr)
+ sys.exit(2)
+
+
+ def die(message):
+- print >>sys.stderr, 'error:', message
++ print('error:', message, file=sys.stderr)
+ sys.exit(2)
+
+
=====================================
debian/patches/do_not_refer_to_non-existing_images.patch
=====================================
@@ -17,12 +17,3 @@ Description: When trying to solve the potential privacy breach issue and
### Input
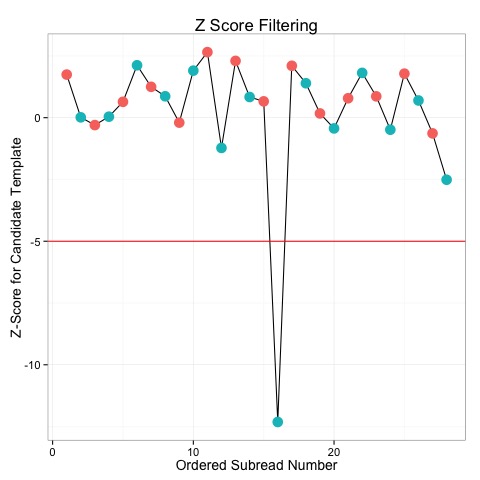
The ccs program needs a .subreads.bam file containing the subreads for each SMRTbell sequenced. Older versions of the PacBio RS software outputted data in bas.h5 files, while the new software outputs BAM files. If you have a bas.h5 file from the older software, you will need to convert it into a BAM. This can be done with the tool bax2bam which simply needs the name of any bas.h5 files to convert and the prefix of the output file. Assuming your original file is named mydata.bas.h5, you can produce a file mynewbam.subreads.bam with the following command.
-@@ -135,8 +131,6 @@ The Z-score for a subread is a metric wh
-
- Subreads with very low Z-scores are very unlikely to have been produced according to the CCS model, and so represent outliers. For example, the plot below shows the Z-scores for several subreads. With a -5 cutoff, we can see that one subread is excluded from the data.
-
--
--
-
- ## CCS Yield Report
-
=====================================
debian/patches/series
=====================================
@@ -2,3 +2,4 @@ add_missing_library_files
git-version.patch
no-static-linking.patch
do_not_refer_to_non-existing_images.patch
+2to3.patch
=====================================
debian/rules
=====================================
@@ -27,13 +27,13 @@ binary-arch: $(docs)
%:
dh $@ \
--package=unanimity \
- --no-package=python-consensuscore2 \
+ --no-package=python3-consensuscore2 \
--buildsystem=cmake \
--builddirectory=build
dh $@ \
- --package=python-consensuscore2 \
+ --package=python3-consensuscore2 \
--no-package=unanimity \
- --with python2 \
+ --with python3 \
--buildsystem=pybuild
# We only want to run the configure step once. If we do not do this,
@@ -56,9 +56,9 @@ config-main:
-Dpbcopper_LIBRARIES=$(pbcopper_LIBRARIES)
touch $@
-override_dh_python2:
- dh_python2
- dh_numpy
+override_dh_python3:
+ dh_python3
+ dh_numpy3
override_dh_auto_clean:
dh_auto_clean
View it on GitLab: https://salsa.debian.org/med-team/unanimity/compare/b81e03b27a36704486f6b662494e0124344789b5...07c8d23df1c301d191517b36b1e6d88a3f5dea82
--
View it on GitLab: https://salsa.debian.org/med-team/unanimity/compare/b81e03b27a36704486f6b662494e0124344789b5...07c8d23df1c301d191517b36b1e6d88a3f5dea82
You're receiving this email because of your account on salsa.debian.org.
-------------- next part --------------
An HTML attachment was scrubbed...
URL: <http://alioth-lists.debian.net/pipermail/debian-med-commit/attachments/20191219/37f98a61/attachment-0001.html>
More information about the debian-med-commit
mailing list